5G5F
 
 | | Crystallographic structure of the Tau class glutathione S-transferase MiGSTU in complex with reduced glutathione. | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTATHIONE, TAU CLASS GLUTATHIONE S-TRANSFERASE | | Authors: | Valenzuela-Chavira, I, Serrano-Posada, H, Lopez-Zavala, A, Hernandez-Paredes, J, Sotelo-Mundo, R. | | Deposit date: | 2016-05-24 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights Into Ligand Binding to a Glutathione S-Transferase from Mango: Structure, Thermodynamics and Kinetics
Biochimie, 135, 2017
|
|
2F8F
 
 | | Crystal structure of the Y10F mutant of the gluathione s-transferase from schistosoma haematobium | | Descriptor: | GLUTATHIONE, Glutathione S-transferase 28 kDa | | Authors: | Gourlay, L.J, Baiocco, P, Angelucci, F, Miele, A.E, Bellelli, A, Brunori, M. | | Deposit date: | 2005-12-02 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the Mechanism of GSH Activation in Schistosoma haematobium Glutathione-S-transferase by Site-directed Mutagenesis and X-ray Crystallography.
J.Mol.Biol., 360, 2006
|
|
2F3M
 
 | | Structure of human GLUTATHIONE S-TRANSFERASE M1A-1A complexed with 1-(S-(GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXADIENATE ANION | | Descriptor: | 1-(S-GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXA-2,5-DIENE, Glutathione S-transferase Mu 1 | | Authors: | Patskovsky, Y, Patskovska, L, Almo, S.C, Listowsky, I. | | Deposit date: | 2005-11-21 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Transition state model and mechanism of nucleophilic aromatic substitution reactions catalyzed by human glutathione S-transferase M1a-1a.
Biochemistry, 45, 2006
|
|
4ZB6
 
 | |
4ZB8
 
 | |
4ZBB
 
 | |
4ZB9
 
 | |
4ZBA
 
 | |
4GCI
 
 | | Crystal structure of glutahtione s-transferase homolog from yersinia pestis, target EFI-501894, with bound glutathione, monoclinic form | | Descriptor: | CHLORIDE ION, GLUTATHIONE, Glutathione S-transferase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-07-30 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of glutahtione s-transferase homolog from yersinia pestis, target EFI-501894, with bound glutathione, monoclinic form
To be Published
|
|
4HJ2
 
 | | Crystal Structure Analysis of GSTA1-1 in complex with chlorambucil | | Descriptor: | Glutathione S-transferase A1, L-gamma-glutamyl-S-(2-{[4-(3-carboxypropyl)phenyl](2-chloroethyl)amino}ethyl)-L-cysteinylglycine | | Authors: | Karpusas, M, Chiniadis, L, Labrou, N.E. | | Deposit date: | 2012-10-12 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The interaction of the chemotherapeutic drug chlorambucil with human glutathione transferase A1-1: kinetic and structural analysis.
Plos One, 8, 2013
|
|
4HI7
 
 | | Crystal structure of glutathione transferase homolog from drosophilia mojavensis, TARGET EFI-501819, with bound glutathione | | Descriptor: | GI20122, GLUTATHIONE | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-10-11 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of glutathione transferase homolog from drosophilia mojavensis, TARGET EFI-501819, with bound glutathione
To be Published
|
|
4HOJ
 
 | | Crystal structure of glutathione transferase homolog from Neisseria Gonorrhoeae, target EFI-501841, with bound glutathione | | Descriptor: | ACETATE ION, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-10-22 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of glutathione transferase homolog from Neisseria Gonorrhoeae, target EFI-501841, with bound glutathione
To be Published
|
|
5A4W
 
 | | AtGSTF2 from Arabidopsis thaliana in complex with quercetrin | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4-oxo-4H-chromen-3-yl 6-deoxy-alpha-L-mannopyranoside, ACETATE ION, GLUTATHIONE S-TRANSFERASE F2 | | Authors: | Ahmad, L, Rylott, E, Bruce, N.C, Edwards, R, Grogan, G. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural evidence for Arabidopsis glutathione transferase AtGSTF2 functioning as a transporter of small organic ligands.
FEBS Open Bio, 7, 2017
|
|
4HZ4
 
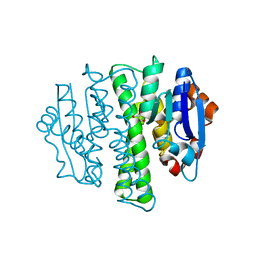 | | Crystal structure of glutathione s-transferase b4xh91 (target efi-501787) from actinobacillus pleuropneumoniae | | Descriptor: | GLYCEROL, Glutathione-S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N.F, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-11-14 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of glutathione s-transferase b4xh91 from Actinobacillus pleuropneumoniae
To be Published
|
|
4ZB7
 
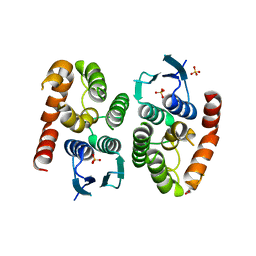 | |
4IKH
 
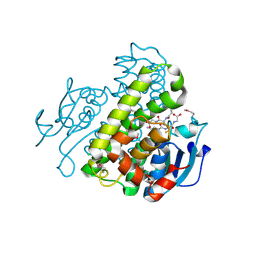 | | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens pf-5, target efi-900003, with two glutathione bound | | Descriptor: | CHLORIDE ION, GLUTATHIONE, Glutathione S-transferase | | Authors: | Vetting, M.W, Sauder, J.M, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Burley, S.K, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-26 | | Release date: | 2013-01-16 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens pf-5, target efi-900003, with two glutathione bound
To be Published
|
|
4ZBD
 
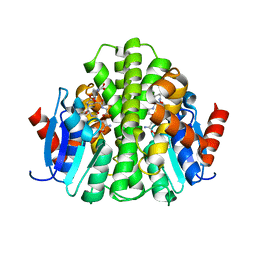 | |
5A4V
 
 | | AtGSTF2 from Arabidopsis thaliana in complex with quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, ACETATE ION, GLUTATHIONE S-TRANSFERASE F2 | | Authors: | Ahmad, L, Rylott, E, Bruce, N.C, Edwards, R, Grogan, G. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural evidence for Arabidopsis glutathione transferase AtGSTF2 functioning as a transporter of small organic ligands.
FEBS Open Bio, 7, 2017
|
|
4GSN
 
 | |
5A4U
 
 | | AtGSTF2 from Arabidopsis thaliana in complex with indole-3-aldehyde | | Descriptor: | 1H-INDOLE-3-CARBALDEHYDE, ACETATE ION, GLUTATHIONE S-TRANSFERASE F2 | | Authors: | Ahmad, L, Rylott, E, Bruce, N.C, Edwards, R, Grogan, G. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for Arabidopsis glutathione transferase AtGSTF2 functioning as a transporter of small organic ligands.
FEBS Open Bio, 7, 2017
|
|
5A34
 
 | | The crystal structure of the GST-like domains complex of EPRS-AIMP2 | | Descriptor: | AMINOACYL TRNA SYNTHASE COMPLEX-INTERACTING MULTIFUNCTIONAL PROTEIN 2, BIFUNCTIONAL GLUTAMATE/PROLINE--TRNA LIGASE, GLYCEROL | | Authors: | Cho, H.Y, Kang, B.S. | | Deposit date: | 2015-05-27 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Assembly of Multi-tRNA Synthetase Complex Via Heterotetrameric Glutathione Transferase-Homology Domains.
J.Biol.Chem., 290, 2015
|
|
5A5H
 
 | |
4I97
 
 | |
4J2F
 
 | | Crystal structure of a glutathione transferase family member from Ricinus communis, target EFI-501866 | | Descriptor: | GLYCEROL, Glutathione s-transferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-02-04 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Ricinus communis, target EFI-501866
TO BE PUBLISHED
|
|
4IVF
 
 | | Crystal structure of glutathione transferase homolog from Lodderomyces elongisporus, target EFI-501753, with two GSH per subunit | | Descriptor: | CITRIC ACID, GLUTATHIONE, Putative uncharacterized protein | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-01-22 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glutathione transferase homolog from Lodderomyces elongisporus, target EFI-501753, with two GSH per subunit
To be Published
|
|
