2DUH
 
 | |
2DUG
 
 | |
2DUI
 
 | |
2I3Y
 
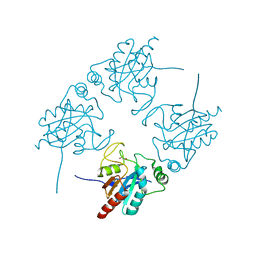 | | Crystal structure of human glutathione peroxidase 5 | | Descriptor: | 1,2-ETHANEDIOL, Epididymal secretory glutathione peroxidase | | Authors: | Kavanagh, K.L, Johansson, C, Rojkova, A, Umeano, C, Bunkoczi, G, Gileadi, O, von Delft, F, Weigelt, J, Arrowsmith, C, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-21 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human glutathione peroxidase 5
To be published
|
|
2I9K
 
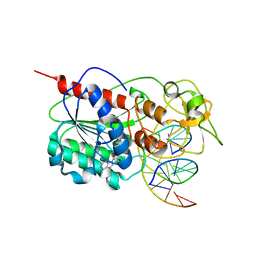 | | Engineered Extrahelical Base Destabilization Enhances Sequence Discrimination of DNA Methyltransferase M.HhaI | | Descriptor: | 5'-D(*T*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3', Modification methylase HhaI, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Youngblood, B, Shieh, F.K, De Los Rios, S, Perona, J.J, Reich, N.O. | | Deposit date: | 2006-09-05 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Engineered Extrahelical Base Destabilization Enhances Sequence Discrimination of DNA Methyltransferase M.HhaI
J.Mol.Biol., 362, 2006
|
|
2IB6
 
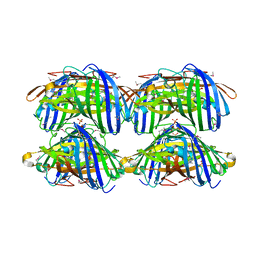 | | Structural characterization of a blue chromoprotein and its yellow mutant from the sea anemone cnidopus japonicus | | Descriptor: | PHOSPHATE ION, Yellow mutant chromo protein | | Authors: | Chan, M.C.Y, Bosanac, I, Ho, D, Prive, G, Ikura, M. | | Deposit date: | 2006-09-10 | | Release date: | 2006-10-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of a Blue Chromoprotein and Its Yellow Mutant from the Sea Anemone Cnidopus Japonicus
J.Biol.Chem., 281, 2006
|
|
2IB5
 
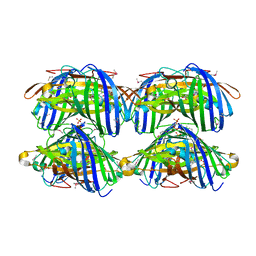 | |
2ICR
 
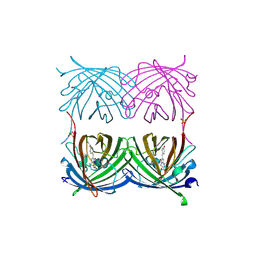 | | Red fluorescent protein zRFP574 from Zoanthus sp. | | Descriptor: | Red fluorescent protein zoanRFP, SULFATE ION | | Authors: | Pletnev, S, Pletneva, N, Tikhonova, T, Pletnev, V. | | Deposit date: | 2006-09-13 | | Release date: | 2007-10-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Refined crystal structures of red and green fluorescent proteins from the button polyp Zoanthus.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2IE2
 
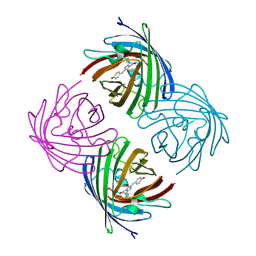 | |
2IOV
 
 | | Bright-state structure of the reversibly switchable fluorescent protein Dronpa | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Stiel, A.C, Trowitzsch, S, Weber, G, Andresen, M, Eggeling, C, Hell, S.W, Jakobs, S, Wahl, M.C. | | Deposit date: | 2006-10-11 | | Release date: | 2006-12-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 A bright-state structure of the reversibly switchable fluorescent protein Dronpa guides the generation of fast switching variants
Biochem.J., 402, 2007
|
|
2NTM
 
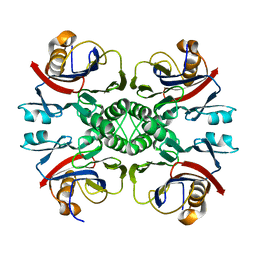 | |
2NTL
 
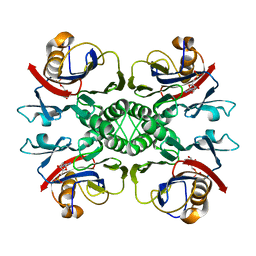 | | Crystal structure of PurO/AICAR from Methanothermobacter thermoautotrophicus | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, IMP cyclohydrolase | | Authors: | Kang, Y.N, Tran, A, White, R.H, Ealick, S.E. | | Deposit date: | 2006-11-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A novel function for the N-terminal nucleophile hydrolase fold demonstrated by the structure of an archaeal inosine monophosphate cyclohydrolase.
Biochemistry, 46, 2007
|
|
2NTK
 
 | | Crystal structure of PurO/IMP from Methanothermobacter thermoautotrophicus | | Descriptor: | IMP cyclohydrolase, INOSINIC ACID | | Authors: | Kang, Y.N, Tran, A, White, R.H, Ealick, S.E. | | Deposit date: | 2006-11-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A novel function for the N-terminal nucleophile hydrolase fold demonstrated by the structure of an archaeal inosine monophosphate cyclohydrolase.
Biochemistry, 46, 2007
|
|
2JAD
 
 | |
2O2B
 
 | |
2O24
 
 | |
2O29
 
 | |
2OGR
 
 | |
2OJK
 
 | | Crystal Structure of Green Fluorescent Protein from Zoanthus sp at 2.2 A Resolution | | Descriptor: | GFP-like fluorescent chromoprotein FP506 | | Authors: | Pletneva, N.V, Pletnev, S.V, Tikhonova, T.V, Pletnev, V.Z. | | Deposit date: | 2007-01-12 | | Release date: | 2007-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined crystal structures of red and green fluorescent proteins from the button polyp Zoanthus.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2JEE
 
 | | Xray structure of E. coli YiiU | | Descriptor: | CELL DIVISION PROTEIN ZAPB | | Authors: | Moller-Jensen, J, Gerdes, K, Lowe, J. | | Deposit date: | 2007-01-16 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel Coiled-Coil Cell Division Factor Zapb Stimulates Z Ring Assembly and Cell Division.
Mol.Microbiol., 68, 2008
|
|
2OKW
 
 | |
2OKY
 
 | |
2ON3
 
 | | A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 3-aminooxy-1-aminopropane | | Descriptor: | 3-AMINOOXY-1-AMINOPROPANE, Ornithine decarboxylase | | Authors: | Dufe, V.T, Ingner, D, Heby, O, Khomutov, A.R, Persson, L, Al-Karadaghi, S. | | Deposit date: | 2007-01-23 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 1-amino-oxy-3-aminopropane.
Biochem.J., 405, 2007
|
|
2OO7
 
 | | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (T179I/Q199R) | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Counago, R, Wilson, C.J, Wu, G, Myers, J.C, Wittung-Stafshede, P, Shamoo, Y. | | Deposit date: | 2007-01-25 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (T179I/Q199R)
To be Published
|
|
2OO0
 
 | | A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 3-aminooxy-1-aminopropane | | Descriptor: | 3-AMINOOXY-1-AMINOPROPANE, ACETATE ION, Ornithine decarboxylase, ... | | Authors: | Dufe, V.T, Ingner, D, Khomutov, A.R, Heby, O, Persson, L, Al-Karadaghi, S. | | Deposit date: | 2007-01-25 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 1-amino-oxy-3-aminopropane.
Biochem.J., 405, 2007
|
|
