1SR3
 
 | |
1SR4
 
 | | Crystal Structure of the Haemophilus ducreyi cytolethal distending toxin | | Descriptor: | BROMIDE ION, Cytolethal distending toxin subunit A, cytolethal distending toxin protein B, ... | | Authors: | Nesic, D, Hsu, Y, Stebbins, C.E. | | Deposit date: | 2004-03-22 | | Release date: | 2004-06-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Assembly and Function of a Bacterial Genotoxin
Nature, 429, 2004
|
|
1SR5
 
 | | ANTITHROMBIN-ANHYDROTHROMBIN-HEPARIN TERNARY COMPLEX STRUCTURE | | Descriptor: | 2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-deoxy-2,3,6-tri-O-methyl-alpha-D-xylo-hexopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfo-alpha-D-glucopyranose-(1-4)-(2R,3R,4S,5S,6S)-6-(dihydroxymethyl)-3,4-dimethoxytetrahydro-2H-pyran-2,5-diol-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-(2R,3R,4S,5S,6R)-6-(dihydroxymethyl)-3,4-dimethoxytetrahydro-2H-pyran-2,5-diol-(1-4)-1,5-anhydro-3-O-methyl-2,6-di-O-sulfo-D-glucitol, ... | | Authors: | Dementiev, A, Petitou, M, Gettins, P.G. | | Deposit date: | 2004-03-22 | | Release date: | 2004-08-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The ternary complex of antithrombin-anhydrothrombin-heparin reveals the basis of inhibitor specificity.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SR6
 
 | | Structure of nucleotide-free scallop myosin S1 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Myosin essential light chain, ... | | Authors: | Risal, D, Gourinath, S, Himmel, D.M, Szent-Gyorgyi, A.G, Cohen, C. | | Deposit date: | 2004-03-22 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Myosin subfragment 1 structures reveal a partially bound nucleotide and a complex salt bridge that helps couple nucleotide and actin binding.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1SR7
 
 | | Progesterone Receptor Hormone Binding Domain with Bound Mometasone Furoate | | Descriptor: | GLYCEROL, MOMETASONE FUROATE, Progesterone receptor, ... | | Authors: | Madauss, K.P, Deng, S.-J, Austin, R.J, Lambert, M.H, McLay, I, Pritchard, J, Short, S.A, Stewart, E.L, Uings, I.J, Williams, S.P. | | Deposit date: | 2004-03-22 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Progesterone receptor ligand binding pocket flexibility: crystal structures of the norethindrone and mometasone furoate complexes
J.Med.Chem., 47, 2004
|
|
1SR8
 
 | | Structural Genomics, 1.9A crystal structure of cobalamin biosynthesis protein (cbiD) from Archaeoglobus fulgidus | | Descriptor: | cobalamin biosynthesis protein (cbiD) | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-03-22 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9A crystal structure of cobalamin biosynthesis protein (cbiD) from Archaeoglobus fulgidus
To be Published
|
|
1SR9
 
 | | Crystal Structure of LeuA from Mycobacterium tuberculosis | | Descriptor: | 2-isopropylmalate synthase, 3-METHYL-2-OXOBUTANOIC ACID, CHLORIDE ION, ... | | Authors: | Koon, N, Squire, C.J, Baker, E.N. | | Deposit date: | 2004-03-22 | | Release date: | 2004-05-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of LeuA from Mycobacterium tuberculosis, a key enzyme in leucine biosynthesis
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1SRA
 
 | | STRUCTURE OF A NOVEL EXTRACELLULAR CA2+-BINDING MODULE IN BM-40(SLASH)SPARC(SLASH)OSTEONECTIN | | Descriptor: | CALCIUM ION, SPARC | | Authors: | Hohenester, E, Maurer, P, Hohenadl, C, Timpl, R, Jansonius, J.N, Engel, J. | | Deposit date: | 1995-08-21 | | Release date: | 1996-03-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a novel extracellular Ca(2+)-binding module in BM-40.
Nat.Struct.Biol., 3, 1996
|
|
1SRB
 
 | | CONFORMATIONAL STUDIES ON SRTB, A NON-SELECTIVE ENDOTHELIN RECEPTOR AGONIST, AND ON IRL 1620, AN ETB RECEPTOR SPECIFIC AGONIST | | Descriptor: | Sarafotoxins precursor | | Authors: | Smith, R, Atkins, A.R. | | Deposit date: | 1994-09-06 | | Release date: | 1994-11-01 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | 1H NMR studies of sarafotoxin SRTb, a nonselective endothelin receptor agonist, and IRL 1620, an ETB receptor-specific agonist.
Biochemistry, 34, 1995
|
|
1SRD
 
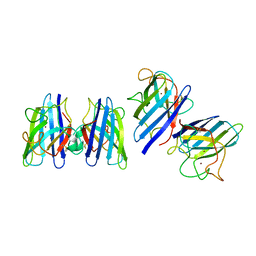 | | Three-dimensional structure of CU,ZN-superoxide dismutase from spinach at 2.0 Angstroms resolution | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Kitagawa, Y, Katsube, Y. | | Deposit date: | 1993-04-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of Cu,Zn-superoxide dismutase from spinach at 2.0 A resolution.
J.Biochem.(Tokyo), 109, 1991
|
|
1SRE
 
 | |
1SRF
 
 | |
1SRG
 
 | |
1SRH
 
 | |
1SRI
 
 | |
1SRJ
 
 | |
1SRK
 
 | | Solution structure of the third zinc finger domain of FOG-1 | | Descriptor: | ZINC ION, Zinc finger protein ZFPM1 | | Authors: | Simpson, R.J.Y, Lee, S.H.Y, Bartle, N, Matthews, J.M, Mackay, J.P, Crossley, M. | | Deposit date: | 2004-03-22 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Classic Zinc Finger from Friend of GATA Mediates an Interaction with the Coiled-coil of Transforming Acidic Coiled-coil 3.
J.Biol.Chem., 279, 2004
|
|
1SRL
 
 | | 1H AND 15N ASSIGNMENTS AND SECONDARY STRUCTURE OF THE SRC SH3 DOMAIN | | Descriptor: | SRC TYROSINE KINASE SH3 DOMAIN | | Authors: | Yu, H, Rosen, M.K, Shin, T.B, Seidel-Dugan, C, Brugge, J.S, Schreiber, S.L. | | Deposit date: | 1994-03-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 1H and 15N assignments and secondary structure of the Src SH3 domain.
FEBS Lett., 324, 1993
|
|
1SRM
 
 | | 1H AND 15N ASSIGNMENTS AND SECONDARY STRUCTURE OF THE SRC SH3 DOMAIN | | Descriptor: | SRC TYROSINE KINASE SH3 DOMAIN | | Authors: | Yu, H, Rosen, M.K, Shin, T.B, Seidel-Dugan, C, Brugge, J.S, Schreiber, S.L. | | Deposit date: | 1994-03-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 1H and 15N assignments and secondary structure of the Src SH3 domain.
FEBS Lett., 324, 1993
|
|
1SRN
 
 | |
1SRO
 
 | | S1 RNA BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | PNPASE | | Authors: | Bycroft, M. | | Deposit date: | 1996-11-27 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold.
Cell(Cambridge,Mass.), 88, 1997
|
|
1SRP
 
 | | STRUCTURAL ANALYSIS OF SERRATIA PROTEASE | | Descriptor: | CALCIUM ION, SERRALYSIN, ZINC ION | | Authors: | Hamada, K, Hiramatsu, H, Katsuya, Y, Hata, Y, Katsube, Y. | | Deposit date: | 1994-11-02 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Serratia protease, a zinc-dependent proteinase from Serratia sp. E-15, containing a beta-sheet coil motif at 2.0 A resolution.
J.Biochem.(Tokyo), 119, 1996
|
|
1SRQ
 
 | | Crystal Structure of the Rap1GAP catalytic domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GTPase-activating protein 1, SULFATE ION | | Authors: | Daumke, O, Weyand, M, Chakrabarti, P.P, Vetter, I.R, Wittinghofer, A. | | Deposit date: | 2004-03-23 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The GTPase activating protein Rap1GAP uses a catalytic asparagine
Nature, 429, 2004
|
|
1SRR
 
 | | CRYSTAL STRUCTURE OF A PHOSPHATASE RESISTANT MUTANT OF SPORULATION RESPONSE REGULATOR SPO0F FROM BACILLUS SUBTILIS | | Descriptor: | CALCIUM ION, SPORULATION RESPONSE REGULATORY PROTEIN | | Authors: | Madhusudan, Whiteley, J.M, Hoch, J.A, Zapf, J, Xuong, N.H, Varughese, K.I. | | Deposit date: | 1996-04-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a phosphatase-resistant mutant of sporulation response regulator Spo0F from Bacillus subtilis.
Structure, 4, 1996
|
|
1SRS
 
 | | SERUM RESPONSE FACTOR (SRF) CORE COMPLEXED WITH SPECIFIC SRE DNA | | Descriptor: | DNA (5'-D(*CP*CP*(5IU)P*TP*CP*CP*TP*AP*AP*TP*TP*AP*GP*GP*CP*CP*AP*TP*G)-3'), DNA (5'-D(*CP*CP*AP*TP*GP*GP*CP*CP*TP*AP*AP*TP*TP*AP*GP*GP*A P*AP*G)-3'), PROTEIN (SERUM RESPONSE FACTOR (SRF)) | | Authors: | Pellegrini, L, Tan, S, Richmond, T.J. | | Deposit date: | 1995-07-28 | | Release date: | 1995-07-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of serum response factor core bound to DNA.
Nature, 376, 1995
|
|
