1SFL
 
 | | 1.9A Crystal structure of Staphylococcus aureus type I 3-dehydroquinase, apo form | | Descriptor: | 3-dehydroquinate dehydratase | | Authors: | Nichols, C.E, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-02-20 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Staphylococcus aureus type I dehydroquinase from enzyme turnover experiments.
Proteins, 56, 2004
|
|
1SFN
 
 | |
1SFO
 
 | | RNA POLYMERASE II STRAND SEPARATED ELONGATION COMPLEX | | Descriptor: | DNA STRAND, DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, ... | | Authors: | Westover, K.D, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2004-02-20 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Structural Basis of Transcription: Separation of RNA from DNA by RNA Polymerase II
Science, 303, 2004
|
|
1SFP
 
 | | CRYSTAL STRUCTURE OF ACIDIC SEMINAL FLUID PROTEIN (ASFP) AT 1.9 A RESOLUTION: A BOVINE POLYPEPTIDE FROM THE SPERMADHESIN FAMILY | | Descriptor: | ASFP | | Authors: | Romao, M.J, Kolln, I, Dias, J.M, Carvalho, A.L, Romero, A, Varela, P.F, Sanz, L, Topfer-Petersen, E, Calvete, J.J. | | Deposit date: | 1997-06-24 | | Release date: | 1998-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of two spermadhesins reveal the CUB domain fold.
Nat.Struct.Biol., 4, 1997
|
|
1SFQ
 
 | | Fast form of thrombin mutant R(77a)A bound to PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, ... | | Authors: | Pineda, A.O, Carrell, C.J, Bush, L.A, Prasad, S, Caccia, S, Chen, Z.W, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-02-20 | | Release date: | 2004-06-08 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular dissection of na+ binding to thrombin.
J.Biol.Chem., 279, 2004
|
|
1SFR
 
 | | Crystal Structure of the Mycobacterium tuberculosis Antigen 85A Protein | | Descriptor: | Antigen 85-A | | Authors: | Ronning, D.R, Vissa, V, Besra, G.S, Belisle, J.T, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-02-20 | | Release date: | 2004-07-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mycobacterium tuberculosis Antigen 85A and 85C Structures Confirm Binding Orientation and Conserved Substrate Specificity
J.Biol.Chem., 279, 2004
|
|
1SFS
 
 | | 1.07 A crystal structure of an uncharacterized B. stearothermophilus protein | | Descriptor: | Hypothetical protein, PHOSPHATE ION | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Moy, S.F, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | 1.07 A crystal structure of an uncharacterized B. stearothermophilus protein
To be Published
|
|
1SFT
 
 | | ALANINE RACEMASE | | Descriptor: | ACETATE ION, Alanine racemase | | Authors: | Shaw, J.P, Petsko, G.A, Ringe, D. | | Deposit date: | 1996-09-20 | | Release date: | 1997-02-12 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of the structure of alanine racemase from Bacillus stearothermophilus at 1.9-A resolution.
Biochemistry, 36, 1997
|
|
1SFU
 
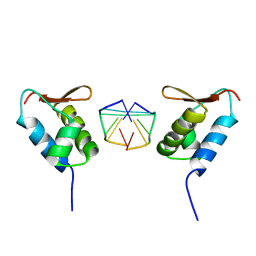 | | Crystal structure of the viral Zalpha domain bound to left-handed Z-DNA | | Descriptor: | 34L protein, 5'-D(*T*CP*GP*CP*GP*CP*G)-3' | | Authors: | Ha, S.C, Van Quyen, D, Wu, C.A, Lowenhaupt, K, Rich, A, Kim, Y.G, Kim, K.K. | | Deposit date: | 2004-02-20 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A poxvirus protein forms a complex with left-handed Z-DNA: crystal structure of a Yatapoxvirus Zalpha bound to DNA.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1SFV
 
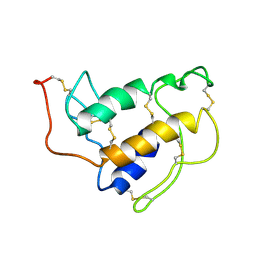 | | PORCINE PANCREAS PHOSPHOLIPASE A2, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Van Den Berg, B, Tessari, M, Boelens, R, Dijkman, R, Kaptein, R, De Haas, G.H, Verheij, H.M. | | Deposit date: | 1996-02-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic phospholipase A2 complexed with micelles and a competitive inhibitor.
J.Biomol.NMR, 5, 1995
|
|
1SFW
 
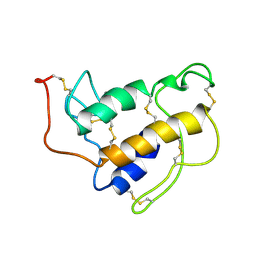 | | PORCINE PANCREAS PHOSPHOLIPASE A2, NMR, 18 STRUCTURES | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Van Den Berg, B, Tessari, M, Boelens, R, Dijkman, R, Kaptein, R, De Haas, G.H, Verheij, H.M. | | Deposit date: | 1996-02-23 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic phospholipase A2 complexed with micelles and a competitive inhibitor.
J.Biomol.NMR, 5, 1995
|
|
1SFX
 
 | | X-ray crystal structure of putative HTH transcription regulator from Archaeoglobus fulgidus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Conserved hypothetical protein AF2008 | | Authors: | Osipiuk, J, Skarina, T, Savchenko, A, Edwards, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystal structure of putative HTH transcription regulator from Archaeoglobus fulgidus
To be Published
|
|
1SFY
 
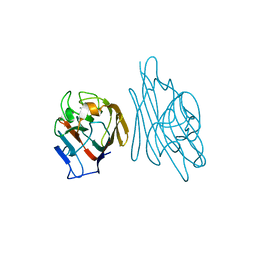 | | Crystal structure of recombinant Erythrina corallodandron Lectin | | Descriptor: | CALCIUM ION, Lectin, MANGANESE (II) ION, ... | | Authors: | Kulkarni, K.A, Srivastava, A, Mitra, N, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2004-02-21 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Effect of glycosylation on the structure of Erythrina corallodendron lectin.
Proteins, 56, 2004
|
|
1SG0
 
 | | Crystal structure analysis of QR2 in complex with resveratrol | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NRH dehydrogenase [quinone] 2, RESVERATROL, ... | | Authors: | Buryanovskyy, L, Fu, Y, Boyd, M, Ma, Y, Tsieh, T.C, Wu, J.M, Zhang, Z. | | Deposit date: | 2004-02-22 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of quinone reductase 2 in complex with resveratrol
Biochemistry, 43, 2004
|
|
1SG1
 
 | |
1SG2
 
 | |
1SG3
 
 | | Structure of allantoicase | | Descriptor: | Allantoicase | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Sorel, I, Graille, M, Meyer, P, Liger, D, Blondeau, K, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2004-02-23 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Yeast Allantoicase Reveals a Repeated Jelly Roll Motif
J.Biol.Chem., 279, 2004
|
|
1SG4
 
 | | Crystal structure of human mitochondrial delta3-delta2-enoyl-CoA isomerase | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, mitochondrial, OCTANOYL-COENZYME A | | Authors: | Partanen, S.T, Novikov, D.K, Popov, A.N, Mursula, A.M, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2004-02-23 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A crystal structure of human mitochondrial Delta3-Delta2-enoyl-CoA isomerase shows a novel mode of binding for the fatty acyl group.
J.Mol.Biol., 342, 2004
|
|
1SG5
 
 | |
1SG6
 
 | | Crystal structure of Aspergillus nidulans 3-dehydroquinate synthase (AnDHQS) in complex with Zn2+ and NAD+, at 1.7D | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pentafunctional AROM polypeptide, ZINC ION | | Authors: | Nichols, C.E, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-02-23 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the 'open' form of Aspergillus nidulans 3-dehydroquinate synthase at 1.7 A resolution from crystals grown following enzyme turnover.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1SG7
 
 | | NMR solution structure of the putative cation transport regulator ChaB | | Descriptor: | Putative Cation transport regulator chaB | | Authors: | Osborne, M.J, Siddiqui, N, Cygler, M, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-02-23 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of ChaB, a putative membrane ion antiporter regulator from Escherichia coli
BMC STRUCT.BIOL., 4, 2004
|
|
1SG8
 
 | | Crystal structure of the procoagulant fast form of thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, thrombin | | Authors: | Pineda, A.O, Carrell, C.J, Bush, L.A, Prasad, S, Caccia, S, Chen, Z.W, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-02-23 | | Release date: | 2004-06-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular dissection of na+ binding to thrombin.
J.Biol.Chem., 279, 2004
|
|
1SG9
 
 | | Crystal structure of Thermotoga maritima protein HEMK, an N5-glutamine methyltransferase | | Descriptor: | GLUTAMINE, S-ADENOSYLMETHIONINE, hemK protein | | Authors: | Agarwal, R, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-02-23 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel mode of dimerization via formation of a glutamate anhydride crosslink in a protein crystal structure.
Proteins, 71, 2008
|
|
1SGC
 
 | |
1SGD
 
 | | ASP 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pH's
To be Published
|
|
