1QNC
 
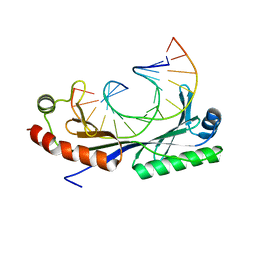 | | Crystal structure of the A(-31) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | Descriptor: | DNA (5'-D(*GP*CP*AP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*TP*TP*TP*AP*TP*TP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | Authors: | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | Deposit date: | 1999-10-14 | | Release date: | 2000-02-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1QND
 
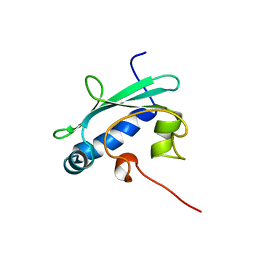 | | STEROL CARRIER PROTEIN-2, NMR, 20 STRUCTURES | | Descriptor: | NONSPECIFIC LIPID-TRANSFER PROTEIN | | Authors: | Lopez-Garcia, F, Szyperski, T, Dyer, J.H, Choinowski, T, Seedorf, U, Hauser, H, Wuthrich, K. | | Deposit date: | 1999-10-14 | | Release date: | 2000-07-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Sterol Carrier Protein-2: Implications for the Biological Role
J.Mol.Biol., 295, 2000
|
|
1QNE
 
 | |
1QNF
 
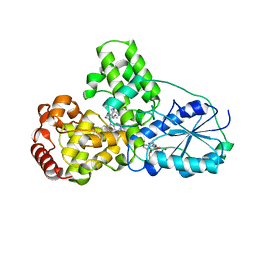 | | STRUCTURE OF PHOTOLYASE | | Descriptor: | 8-HYDROXY-10-(D-RIBO-2,3,4,5-TETRAHYDROXYPENTYL)-5-DEAZAISOALLOXAZINE, FLAVIN-ADENINE DINUCLEOTIDE, PHOTOLYASE | | Authors: | Miki, K, Kitadokoro, K. | | Deposit date: | 1997-07-04 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of DNA photolyase from Anacystis nidulans
Nat.Struct.Biol., 4, 1997
|
|
1QNG
 
 | |
1QNH
 
 | |
1QNI
 
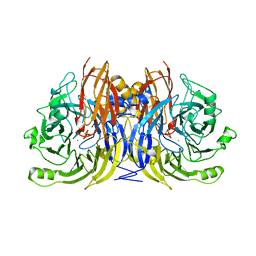 | | Crystal Structure of Nitrous Oxide Reductase from Pseudomonas nautica, at 2.4A Resolution | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Brown, K, Tegoni, M, Cambillau, C. | | Deposit date: | 1999-10-15 | | Release date: | 2000-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel type of catalytic copper cluster in nitrous oxide reductase.
Nat.Struct.Biol., 7, 2000
|
|
1QNJ
 
 | | THE STRUCTURE OF NATIVE PORCINE PANCREATIC ELASTASE AT ATOMIC RESOLUTION (1.1 A) | | Descriptor: | ELASTASE, SODIUM ION, SULFATE ION | | Authors: | Wurtele, M, Hahn, M, Hilpert, K, Hohne, W. | | Deposit date: | 1999-10-15 | | Release date: | 2000-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Structure of Native Porcine Pancreatic Elastase at 1.1 A
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QNK
 
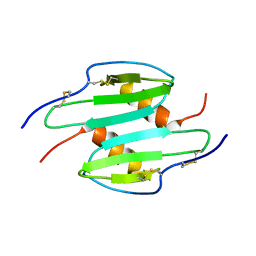 | | TRUNCATED HUMAN GROB[5-73], NMR, 20 STRUCTURES | | Descriptor: | C-X-C motif chemokine 2 | | Authors: | Qian, Y.Q, Johanson, K, McDevitt, P. | | Deposit date: | 1999-10-18 | | Release date: | 2000-02-04 | | Last modified: | 2018-06-13 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of truncated human GRObeta [5-73] and its structural comparison with CXC chemokine family members GROalpha and IL-8.
J. Mol. Biol., 294, 1999
|
|
1QNL
 
 | |
1QNM
 
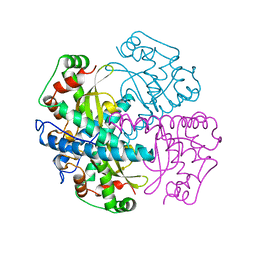 | | HUMAN MANGANESE SUPEROXIDE DISMUTASE MUTANT Q143N | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Guan, Y, Tainer, J.A. | | Deposit date: | 1997-07-03 | | Release date: | 1998-01-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the active site of human manganese superoxide dismutase: the role of glutamine 143.
Biochemistry, 37, 1998
|
|
1QNN
 
 | |
1QNO
 
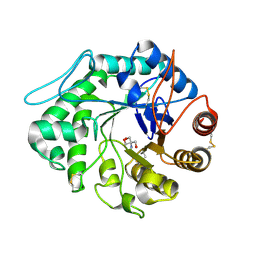 | | The 3-D structure of a Trichoderma reesei b-mannanase from glycoside hydrolase family 5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-1,4-B-D-MANNANASE | | Authors: | Sabini, E, Schubert, H, Murshudov, G, Wilson, K.S, Siika-Aho, M, Penttila, M. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Three-Dimensional Structure of a Trichoderma Reesei Beta-Mannanase from Glycoside Hydrolase Family 5.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QNP
 
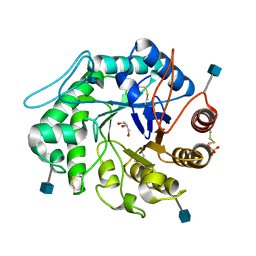 | | The 3-D structure of a Trichoderma reesei b-mannanase from glycoside hydrolase family 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-1,4-B-D-MANNANASE, GLYCEROL, ... | | Authors: | Sabini, E, Schubert, H, Murshudov, G, Wilson, K.S, Siika-Aho, M, Penttila, M. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Three-Dimensional Structure of a Trichoderma Reesei Beta-Mannanase from Glycoside Hydrolase Family 5.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QNQ
 
 | | The 3-D structure of a Trichoderma reesei b-mannanase from glycoside hydrolase family 5 | | Descriptor: | 2,2':6',2''-TERPYRIDINE PLATINUM(II) Chloride, 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-1,4-B-D-MANNANASE, ... | | Authors: | Sabini, E, Schubert, H, Murshudov, G, Wilson, K.S, Siika-Aho, M, Penttila, M. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Three-Dimensional Structure of a Trichoderma Reesei Beta-Mannanase from Glycoside Hydrolase Family 5.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QNR
 
 | | The 3-D structure of a Trichoderma reesei b-mannanase from glycoside hydrolase family 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-1,4-B-D-MANNANASE, GLYCEROL, ... | | Authors: | Sabini, E, Schubert, H, Murshudov, G, Wilson, K.S, Siika-Aho, M, Penttila, M. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Three-Dimensional Structure of a Trichoderma Reesei Beta-Mannanase from Glycoside Hydrolase Family 5.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QNS
 
 | | The 3-D structure of a Trichoderma reesei b-mannanase from glycoside hydrolase family 5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-1,4-B-D-MANNANASE, ... | | Authors: | Sabini, E, Schubert, H, Murshudov, G, Wilson, K.S, Siika-Aho, M, Penttila, M. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Three-Dimensional Structure of a Trichoderma Reesei Beta-Mannanase from Glycoside Hydrolase Family 5
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QNT
 
 | |
1QNU
 
 | | Shiga-Like Toxin I B Subunit Complexed with the Bridged-Starfish Inhibitor | | Descriptor: | ETHYL-CARBAMIC ACID METHYL ESTER, METHYL-CARBAMIC ACID ETHYL ESTER, Shiga toxin 1 variant B subunit, ... | | Authors: | Pannu, N.S, Hayakawa, K, Read, R.J. | | Deposit date: | 1999-10-21 | | Release date: | 2000-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Shiga-like toxins are neutralized by tailored multivalent carbohydrate ligands.
Nature, 403, 2000
|
|
1QNV
 
 | | yeast 5-aminolaevulinic acid dehydratase Lead (Pb) complex | | Descriptor: | 5-AMINOLAEVULINIC ACID DEHYDRATASE, LEAD (II) ION | | Authors: | Erskine, P.T, Senior, N.M, Warren, M.J, Wood, S.P, Cooper, J.B. | | Deposit date: | 1999-10-21 | | Release date: | 2000-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MAD Analyses of Yeast 5-Aminolaevulinic Acid Dehydratase. Their Use in Structure Determination and in Defining the Metal Binding Sites
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QNW
 
 | | lectin II from Ulex europaeus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHITIN BINDING LECTIN, ... | | Authors: | Loris, R, De Greve, H, Dao-Thi, M.-H, Messens, J, Imberty, A, Wyns, L. | | Deposit date: | 1999-10-22 | | Release date: | 1999-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by Lectin II from Ulex Europaeus, a Protein with a Promiscuous Carbohydrate Binding Site
J.Mol.Biol., 301, 2000
|
|
1QNX
 
 | | Ves v 5, an allergen from Vespula vulgaris venom | | Descriptor: | SODIUM ION, VES V 5 | | Authors: | Henriksen, A, Gajhede, M, Spangfort, M.D. | | Deposit date: | 1999-10-25 | | Release date: | 2000-10-26 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Major Venom Allergen of Yellow Jackets, Ves V 5: Structural Characterization of a Pathogenesis-Related Protein Superfamily.
Proteins: Struct.,Funct., Genet., 45, 2001
|
|
1QNY
 
 | | X-ray refinement of D2O soaked crystal of concanavalin A | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION | | Authors: | Habash, J, Raftery, J, Nuttall, R, Price, H.J, Lehmann, M.S, Wilkinson, C, Kalb(Gilboa), A.J, Helliwell, J.R. | | Deposit date: | 1999-10-26 | | Release date: | 2000-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct Determination of the Positions of Deuterium Atoms of Bound Water in Concanavalin a by Neutron Laue Crystallography
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QNZ
 
 | | NMR structure of the 0.5b anti-HIV antibody complex with the gp120 V3 peptide | | Descriptor: | 0.5B ANTIBODY (HEAVY CHAIN), 0.5B ANTIBODY (LIGHT CHAIN), GP120 | | Authors: | Tugarinov, V, Zvi, A, Levy, R, Hayek, Y, Matsushita, S, Anglister, J. | | Deposit date: | 1999-10-26 | | Release date: | 2000-06-03 | | Last modified: | 2018-01-17 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of an Anti-Gp120 Antibody Complex with a V3 Peptide Reveals a Surface Important for Co-Receptor Binding
Structure, 8, 2000
|
|
1QO0
 
 | |
