1K7V
 
 | | Crystal Structure Analysis of crosslinked-WGA3/GlcNAcbeta1,6Galbeta1,4Glc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, agglutinin isolectin 3 | | Authors: | Muraki, M, Ishimura, M, Harata, K. | | Deposit date: | 2001-10-22 | | Release date: | 2002-04-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions of wheat-germ agglutinin with GlcNAc beta 1,6Gal sequence
Biochim.Biophys.Acta, 1569, 2002
|
|
1K7W
 
 | | Crystal Structure of S283A Duck Delta 2 Crystallin Mutant | | Descriptor: | ARGININOSUCCINATE, delta 2 crystallin | | Authors: | Sampaleanu, L.M, Yu, B, Howell, P.L. | | Deposit date: | 2001-10-22 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Mutational analysis of duck delta 2 crystallin and the structure of an inactive mutant with bound substrate provide insight into the enzymatic mechanism of argininosuccinate lyase.
J.Biol.Chem., 277, 2002
|
|
1K7X
 
 | | CRYSTAL STRUCTURE OF THE BETA-SER178PRO MUTANT OF TRYPTOPHAN SYNTHASE | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-22 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the beta Ser178--> Pro mutant of tryptophan synthase. A "knock-out" allosteric enzyme.
J.Biol.Chem., 277, 2002
|
|
1K7Y
 
 | | E. coli MetH C-terminal fragment (649-1227) | | Descriptor: | COBALAMIN, SULFATE ION, methionine synthase | | Authors: | Bandarian, V, Pattridge, K.A, Lennon, B.W, Huddler, D.P, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2001-10-22 | | Release date: | 2001-12-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Domain alternation switches B(12)-dependent methionine synthase to the activation conformation.
Nat.Struct.Biol., 9, 2002
|
|
1K81
 
 | |
1K82
 
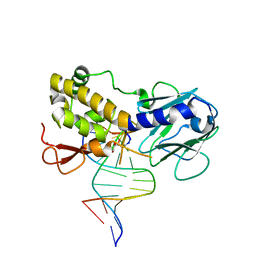 | | Crystal structure of E.coli formamidopyrimidine-DNA glycosylase (Fpg) covalently trapped with DNA | | Descriptor: | 5'-D(*CP*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*TP*TP*CP*CP*TP*CP*CP*TP*GP*G)-3', ZINC ION, ... | | Authors: | Gilboa, R, Zharkov, D.O, Golan, G, Fernandes, A.S, Gerchman, S.E, Matz, E, Kycia, J.H, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-10-22 | | Release date: | 2002-06-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA.
J.Biol.Chem., 277, 2002
|
|
1K83
 
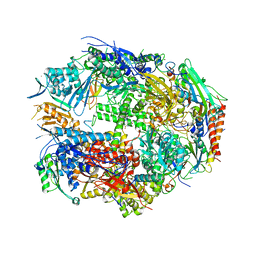 | | Crystal Structure of Yeast RNA Polymerase II Complexed with the Inhibitor Alpha Amanitin | | Descriptor: | ALPHA AMANITIN, DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.2KD POLYPEPTIDE, ... | | Authors: | Bushnell, D.A, Cramer, P, Kornberg, R.D. | | Deposit date: | 2001-10-22 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Transcription: Alpha-Amanitin-RNA Polymerase II Cocrystal at 2.8 A Resolution.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1K85
 
 | | Solution structure of the fibronectin type III domain from Bacillus circulans WL-12 Chitinase A1. | | Descriptor: | CHITINASE A1 | | Authors: | Jee, J.G, Ikegami, T, Hashimoto, M, Kawabata, T, Ikeguchi, M, Watanabe, T, Shirakawa, M. | | Deposit date: | 2001-10-23 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Fibronectin Type III Domain
from Bacillus circulans WL-12 Chitinase A1
J.Biol.Chem., 277, 2002
|
|
1K86
 
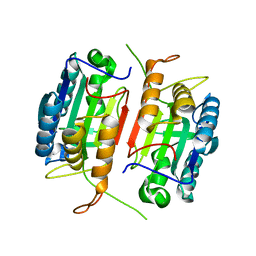 | | Crystal structure of caspase-7 | | Descriptor: | caspase-7 | | Authors: | Chai, J, Wu, Q, Shiozaki, E, Srinivasa, S.M, Alnemri, E.S, Shi, Y. | | Deposit date: | 2001-10-23 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a procaspase-7 zymogen: mechanisms of activation and substrate binding
Cell(Cambridge,Mass.), 107, 2001
|
|
1K88
 
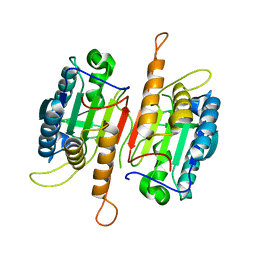 | | Crystal structure of procaspase-7 | | Descriptor: | procaspase-7 | | Authors: | Chai, J, Wu, Q, Shiozaki, E, Srinivasa, S.M, Alnemri, E.S, Shi, Y. | | Deposit date: | 2001-10-23 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a procaspase-7 zymogen: mechanisms of activation and substrate binding
Cell(Cambridge,Mass.), 107, 2001
|
|
1K89
 
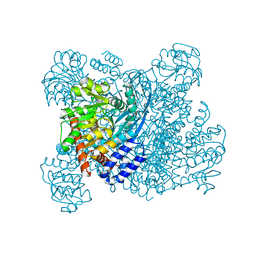 | | K89L MUTANT OF GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE | | Authors: | Stillman, T.J, Migueis, A.M.B, Wang, X.G, Baker, P.J, Britton, K.L, Engel, P.C, Rice, D.W. | | Deposit date: | 1998-06-05 | | Release date: | 1999-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Insights into the mechanism of domain closure and substrate specificity of glutamate dehydrogenase from Clostridium symbiosum.
J.Mol.Biol., 285, 1999
|
|
1K8A
 
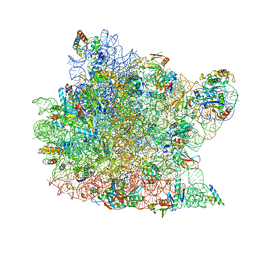 | | Co-crystal structure of Carbomycin A bound to the 50S ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T. | | Deposit date: | 2001-10-23 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
1K8B
 
 | |
1K8C
 
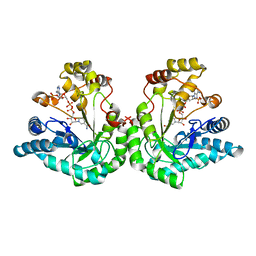 | | Crystal structure of dimeric xylose reductase in complex with NADP(H) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, xylose reductase | | Authors: | Kavanagh, K.L, Klimacek, M, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2001-10-23 | | Release date: | 2002-07-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of apo and holo forms of xylose reductase, a dimeric aldo-keto reductase from Candida tenuis.
Biochemistry, 41, 2002
|
|
1K8D
 
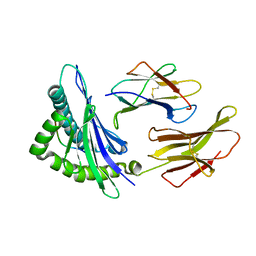 | | crystal structure of the non-classical MHC class Ib Qa-2 complexed with a self peptide | | Descriptor: | 60S RIBOSOMAL PROTEIN, BETA-2-MICROGLOBULIN, QA-2 antigen | | Authors: | He, X, Tabaczewski, P, Ho, J, Stroynowski, I, Garcia, K.C. | | Deposit date: | 2001-10-23 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Promiscuous antigen presentation by the nonclassical MHC Ib Qa-2 is enabled by a shallow, hydrophobic groove and self-stabilized peptide conformation.
Structure, 9, 2001
|
|
1K8F
 
 | | CRYSTAL STRUCTURE OF THE HUMAN C-TERMINAL CAP1-ADENYLYL CYCLASE ASSOCIATED PROTEIN | | Descriptor: | ADENYLYL CYCLASE-ASSOCIATED PROTEIN | | Authors: | Patskovsky, Y.V, Chance, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-10-24 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the actin binding domain of the cyclase-associated protein.
Biochemistry, 43, 2004
|
|
1K8G
 
 | |
1K8H
 
 | |
1K8I
 
 | | CRYSTAL STRUCTURE OF MOUSE H2-DM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC class II H2-M alpha chain, MHC class II H2-M beta 2 chain | | Authors: | Fremont, D.H, Crawford, F, Marrack, P, Hendrickson, W, Kappler, J. | | Deposit date: | 2001-10-24 | | Release date: | 2001-12-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of mouse H2-M.
Immunity, 9, 1998
|
|
1K8J
 
 | | NMR STRUCTURE OF THE CK14 DNA DUPLEX: A PORTION OF THE KNOWN NF-kB SEQUENCE CK1 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1K8K
 
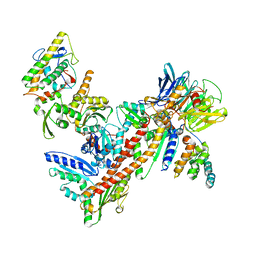 | | Crystal Structure of Arp2/3 Complex | | Descriptor: | ACTIN-LIKE PROTEIN 2, ACTIN-LIKE PROTEIN 3, ARP2/3 COMPLEX 16 KDA SUBUNIT, ... | | Authors: | Robinson, R.C, Turbedsky, K, Kaiser, D.A, Higgs, H.N, Marchand, J.-B, Choe, S, Pollard, T.D. | | Deposit date: | 2001-10-24 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Arp2/3 Complex
Science, 294, 2001
|
|
1K8L
 
 | | XBY6: An analog of CK14 containing 6 dithiophosphate groups | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1K8M
 
 | | Solution Structure of the Lipoic Acid-Bearing Domain of the E2 component of Human, Mitochondrial Branched-Chain alpha-Ketoacid Dehydrogenase | | Descriptor: | E2 component of Branched-Chain alpha-Ketoacid Dehydrogenase | | Authors: | Chang, C.-F, Chou, H.-T, Chuang, J.L, Chuang, D.T, Huang, T.-h. | | Deposit date: | 2001-10-24 | | Release date: | 2001-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the lipoic acid-bearing domain of human mitochondrial branched-chain alpha-keto acid dehydrogenase complex
J.Biol.Chem., 277, 2002
|
|
1K8N
 
 | | NMR structure of the XBY2 DNA duplex, an analog of CK14 containing phosphorodithioate groups at C22 and C24 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1K8O
 
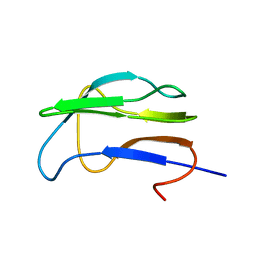 | | Solution Structure of the Lipoic Acid-Bearing Domain of the E2 component of Human, Mitochondrial Branched-Chain alpha-Ketoacid Dehydrogenase | | Descriptor: | E2 component of Branched-Chain alpha-Ketoacid Dehydrogenase | | Authors: | Chang, C.-F, Chou, H.-T, Chuang, J.L, Chuang, D.T, Huang, T.-h. | | Deposit date: | 2001-10-24 | | Release date: | 2001-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the lipoic acid-bearing domain of human mitochondrial branched-chain alpha-keto acid dehydrogenase complex
J.Biol.Chem., 277, 2002
|
|
