1ICL
 
 | |
1ICM
 
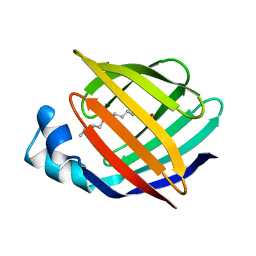 | | ESCHERICHIA COLI-DERIVED RAT INTESTINAL FATTY ACID BINDING PROTEIN WITH BOUND MYRISTATE AT 1.5 A RESOLUTION AND I-FABPARG106-->GLN WITH BOUND OLEATE AT 1.74 A RESOLUTION | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN, MYRISTIC ACID | | Authors: | Eads, J.C, Sacchettini, J.C, Kromminga, A, Gordon, J.I. | | Deposit date: | 1993-09-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Escherichia coli-derived rat intestinal fatty acid binding protein with bound myristate at 1.5 A resolution and I-FABPArg106-->Gln with bound oleate at 1.74 A resolution.
J.Biol.Chem., 268, 1993
|
|
1ICN
 
 | | ESCHERICHIA COLI-DERIVED RAT INTESTINAL FATTY ACID BINDING PROTEIN WITH BOUND MYRISTATE AT 1.5 A RESOLUTION AND I-FABPARG106-->GLN WITH BOUND OLEATE AT 1.74 A RESOLUTION | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN, OLEIC ACID | | Authors: | Eads, J.C, Sacchettini, J.C, Kromminga, A, Gordon, J.I. | | Deposit date: | 1993-09-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Escherichia coli-derived rat intestinal fatty acid binding protein with bound myristate at 1.5 A resolution and I-FABPArg106-->Gln with bound oleate at 1.74 A resolution.
J.Biol.Chem., 268, 1993
|
|
1ICO
 
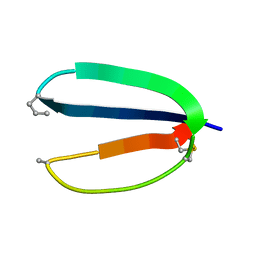 | |
1ICP
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO COMPLEXED WITH PEG400 | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
1ICQ
 
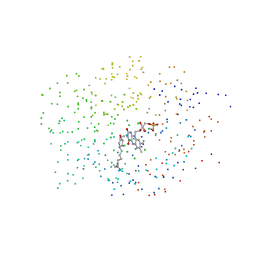 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO COMPLEXED WITH 9R,13R-OPDA | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, 9R,13R-12-OXOPHYTODIENOIC ACID, FLAVIN MONONUCLEOTIDE | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
1ICR
 
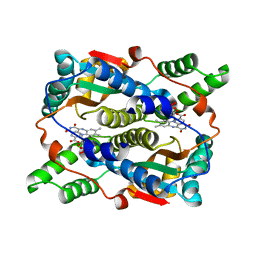 | | THE STRUCTURE OF ESCHERICHIA COLI NITROREDUCTASE COMPLEXED WITH NICOTINIC ACID | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINIC ACID, OXYGEN-INSENSITIVE NAD(P)H NITROREDUCTASE | | Authors: | Lovering, A.L, Hyde, E.I, Searle, P.F, White, S.A. | | Deposit date: | 2001-04-02 | | Release date: | 2001-04-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of Escherichia coli nitroreductase complexed with nicotinic acid: three crystal forms at 1.7 A, 1.8 A and 2.4 A resolution.
J.Mol.Biol., 309, 2001
|
|
1ICS
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, FLAVIN MONONUCLEOTIDE | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
1ICT
 
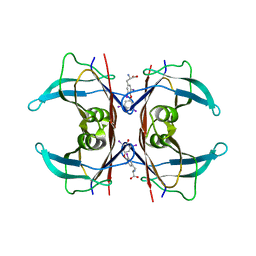 | | MONOCLINIC FORM OF HUMAN TRANSTHYRETIN COMPLEXED WITH THYROXINE (T4) | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, TRANSTHYRETIN | | Authors: | Wojtczak, A, Neumann, P, Cody, V. | | Deposit date: | 2001-04-02 | | Release date: | 2002-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a new polymorphic monoclinic form of human transthyretin at 3 A resolution reveals a mixed complex between unliganded and T4-bound tetramers of TTR.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1ICU
 
 | | THE STRUCTURE OF ESCHERICHIA COLI NITROREDUCTASE COMPLEXED WITH NICOTINIC ACID | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINIC ACID, OXYGEN-INSENSITIVE NAD(P)H NITROREDUCTASE | | Authors: | Lovering, A.L, Hyde, E.I, Searle, P.F, White, S.A. | | Deposit date: | 2001-04-02 | | Release date: | 2001-04-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Escherichia coli nitroreductase complexed with nicotinic acid: three crystal forms at 1.7 A, 1.8 A and 2.4 A resolution.
J.Mol.Biol., 309, 2001
|
|
1ICV
 
 | | THE STRUCTURE OF ESCHERICHIA COLI NITROREDUCTASE COMPLEXED WITH NICOTINIC ACID | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINIC ACID, OXYGEN-INSENSITIVE NAD(P)H NITROREDUCTASE | | Authors: | Lovering, A.L, Hyde, E.I, Searle, P.F, White, S.A. | | Deposit date: | 2001-04-02 | | Release date: | 2001-04-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of Escherichia coli nitroreductase complexed with nicotinic acid: three crystal forms at 1.7 A, 1.8 A and 2.4 A resolution.
J.Mol.Biol., 309, 2001
|
|
1ICW
 
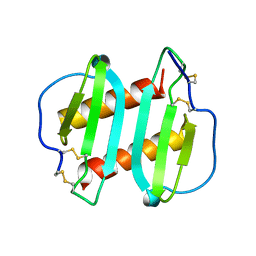 | | INTERLEUKIN-8, MUTANT WITH GLU 38 REPLACED BY CYS AND CYS 50 REPLACED BY ALA | | Descriptor: | INTERLEUKIN-8 | | Authors: | Eigenbrot, C, Lowman, H.B, Chee, L, Artis, D.R. | | Deposit date: | 1996-09-18 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural change and receptor binding in a chemokine mutant with a rearranged disulfide: X-ray structure of E38C/C50AIL-8 at 2 A resolution.
Proteins, 27, 1997
|
|
1ICX
 
 | |
1ICY
 
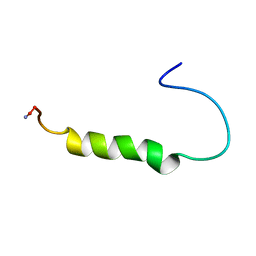 | | [ALA31,PRO32]-PNPY BOUND TO DPC MICELLES | | Descriptor: | NEUROPEPTIDE Y | | Authors: | Bader, R, Rytz, G, Lerch, M, Beck-Sickinger, A.G, Zerbe, O. | | Deposit date: | 2001-04-02 | | Release date: | 2002-05-08 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Key motif to gain selectivity at the neuropeptide Y5-receptor: structure and dynamics of micelle-bound [Ala31, Pro32]-NPY.
Biochemistry, 41, 2002
|
|
1ID0
 
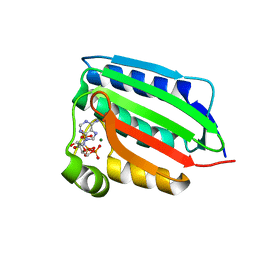 | | CRYSTAL STRUCTURE OF THE NUCLEOTIDE BOND CONFORMATION OF PHOQ KINASE DOMAIN | | Descriptor: | MAGNESIUM ION, PHOQ HISTIDINE KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Marina, A, Mott, C, Auyzenberg, A, Waldburger, C.D, Hendrickson, W.A. | | Deposit date: | 2001-04-02 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and mutational analysis of the PhoQ histidine kinase catalytic domain. Insight into the reaction mechanism.
J.Biol.Chem., 276, 2001
|
|
1ID1
 
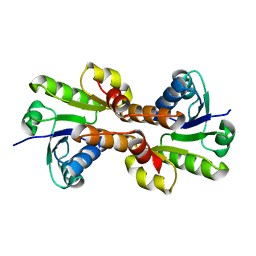 | | CRYSTAL STRUCTURE OF THE RCK DOMAIN FROM E.COLI POTASSIUM CHANNEL | | Descriptor: | PUTATIVE POTASSIUM CHANNEL PROTEIN | | Authors: | Jiang, Y, Pico, A, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2001-04-02 | | Release date: | 2001-04-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RCK domain from the E. coli K+ channel and demonstration of its presence in the human BK channel.
Neuron, 29, 2001
|
|
1ID2
 
 | | CRYSTAL STRUCTURE OF AMICYANIN FROM PARACOCCUS VERSUTUS (THIOBACILLUS VERSUTUS) | | Descriptor: | AMICYANIN, COPPER (II) ION | | Authors: | Romero, A, Nar, H, Messerschmidt, A. | | Deposit date: | 2001-04-03 | | Release date: | 2001-04-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure analysis and refinement at 2.15 A resolution of amicyanin, a type I blue copper protein, from Thiobacillus versutus.
J.Mol.Biol., 236, 1994
|
|
1ID3
 
 | | CRYSTAL STRUCTURE OF THE YEAST NUCLEOSOME CORE PARTICLE REVEALS FUNDAMENTAL DIFFERENCES IN INTER-NUCLEOSOME INTERACTIONS | | Descriptor: | HISTONE H2A.1, HISTONE H2B.2, HISTONE H3, ... | | Authors: | White, C.L, Suto, R.K, Luger, K. | | Deposit date: | 2001-04-03 | | Release date: | 2001-09-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions.
EMBO J., 20, 2001
|
|
1ID4
 
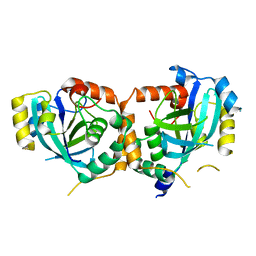 | | CRYSTAL STRUCTURE OF THE CATALYTIC SITE MUTANT (H157Q) OF THE HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | CAPSID PROTEIN P40: ASSEMBLIN PROTEASE | | Authors: | Khayat, R, Batra, R, Massariol, M.J, Lagace, L, Tong, L. | | Deposit date: | 2001-04-03 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Investigating the role of histidine 157 in the catalytic activity of human cytomegalovirus protease.
Biochemistry, 40, 2001
|
|
1ID5
 
 | |
1ID6
 
 | | SOLUTION STRUCTURES OF SYR6 | | Descriptor: | SYR6 | | Authors: | Sato, A, Kawaguchi, K, Kimura, K, Tanimura, R, Sone, S. | | Deposit date: | 2001-04-04 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A peptide mimetic of IFN, the first proof of a small peptidic agonist for heterodimeric cytokine receptor
To be Published
|
|
1ID7
 
 | | SOLUTION STRUCTURE OF SYR6 | | Descriptor: | SYR6 | | Authors: | Sato, A, Kawaguchi, K, Kimura, K, Tanimura, R, Sone, S. | | Deposit date: | 2001-04-04 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A peptide mimetic of IFN, the first proof of a small peptidic agonist for heterodimeric cytokine receptor
To be Published
|
|
1ID8
 
 | | NMR STRUCTURE OF GLUTAMATE MUTASE (B12-BINDING SUBUNIT) COMPLEXED WITH THE VITAMIN B12 NUCLEOTIDE | | Descriptor: | 2-HYDROXY-PROPYL-AMMONIUM, METHYLASPARTATE MUTASE S CHAIN, PHOSPHORIC ACID MONO-[5-(5,6-DIMETHYL-BENZOIMIDAZOL-1-YL)-4-HYDROXY-2-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-YL] ESTER | | Authors: | Tollinger, M, Eichmuller, C, Konrat, R, Huhta, M.S, Marsh, E.N.G, Krautler, B. | | Deposit date: | 2001-04-04 | | Release date: | 2001-06-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The B(12)-binding subunit of glutamate mutase from Clostridium tetanomorphum traps the nucleotide moiety of coenzyme B(12).
J.Mol.Biol., 309, 2001
|
|
1ID9
 
 | | STRUCTURE OF THE HYBRID RNA/DNA R-GCUUCGGC-D[F]U IN PRESENCE OF RH(NH3)6+++ | | Descriptor: | 5'-R(*GP*CP*UP*UP*CP*GP*GP*C)-D(P*(UFP))-3', CHLORIDE ION, RHODIUM HEXAMINE ION | | Authors: | Cruse, W, Saludjian, P, Neuman, A, Prange, T. | | Deposit date: | 2001-04-04 | | Release date: | 2001-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Destabilizing effect of a fluorouracil extra base in a hybrid RNA duplex compared with bromo and chloro analogues.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IDA
 
 | |
