1FKK
 
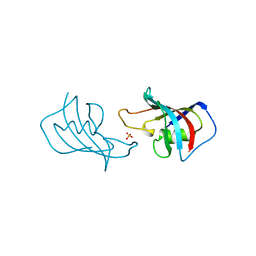 | | ATOMIC STRUCTURE OF FKBP12, AN IMMUNOPHILIN BINDING PROTEIN | | Descriptor: | FK506 BINDING PROTEIN, SULFATE ION | | Authors: | Wilson, K.P, Sintchak, M.D, Thomson, J.A, Navia, M.A. | | Deposit date: | 1995-08-18 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparative X-ray structures of the major binding protein for the immunosuppressant FK506 (tacrolimus) in unliganded form and in complex with FK506 and rapamycin.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1FKL
 
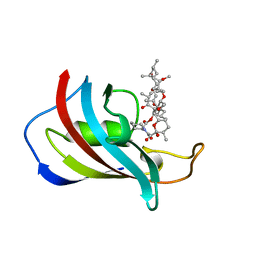 | | ATOMIC STRUCTURE OF FKBP12-RAPAYMYCIN, AN IMMUNOPHILIN-IMMUNOSUPPRESSANT COMPLEX | | Descriptor: | FK506 BINDING PROTEIN, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Wilson, K.P, Sintchak, M.D, Thomson, J.A, Navia, M.A. | | Deposit date: | 1995-08-18 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparative X-ray structures of the major binding protein for the immunosuppressant FK506 (tacrolimus) in unliganded form and in complex with FK506 and rapamycin.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1FKM
 
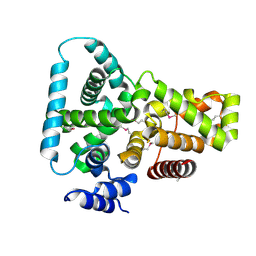 | | CRYSTAL STRUCTURE OF THE YPT/RAB-GAP DOMAIN OF GYP1P | | Descriptor: | PROTEIN (GYP1P) | | Authors: | Rak, A, Fedorov, R, Alexandrov, K, Albert, S, Goody, R.S, Gallwitz, D, Scheidig, A.J. | | Deposit date: | 2000-08-09 | | Release date: | 2001-02-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the GAP domain of Gyp1p: first insights into interaction with Ypt/Rab proteins.
EMBO J., 19, 2000
|
|
1FKN
 
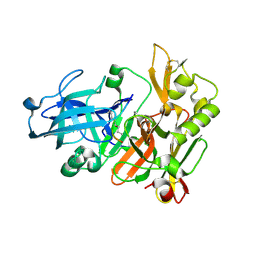 | | Structure of Beta-Secretase Complexed with Inhibitor | | Descriptor: | MEMAPSIN 2, inhibitor | | Authors: | Hong, L, Koelsch, G, Lin, X, Wu, S, Terzyan, S, Ghosh, A, Zhang, X.C, Tang, J. | | Deposit date: | 2000-08-09 | | Release date: | 2000-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the protease domain of memapsin 2 (beta-secretase) complexed with inhibitor.
Science, 290, 2000
|
|
1FKO
 
 | | CRYSTAL STRUCTURE OF NNRTI RESISTANT K103N MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DMP-266(EFAVIRENZ) | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Milton, J, Weaver, K.L, Short, S.A, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-08-10 | | Release date: | 2000-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase.
Structure Fold.Des., 8, 2000
|
|
1FKP
 
 | | CRYSTAL STRUCTURE OF NNRTI RESISTANT K103N MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Milton, J, Weaver, K.L, Short, S.A, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-08-10 | | Release date: | 2000-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase.
Structure Fold.Des., 8, 2000
|
|
1FKQ
 
 | | RECOMBINANT GOAT ALPHA-LACTALBUMIN T29V | | Descriptor: | ALPHA-LACTALBUMIN, CALCIUM ION | | Authors: | Horii, K, Matsushima, M, Tsumoto, K, Kumagai, I. | | Deposit date: | 2000-08-10 | | Release date: | 2001-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of Thr29 to the thermodynamic stability of goat alpha-lactalbumin as determined by experimental and theoretical approaches.
Proteins, 45, 2001
|
|
1FKR
 
 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | Descriptor: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | Authors: | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | Deposit date: | 1992-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
1FKS
 
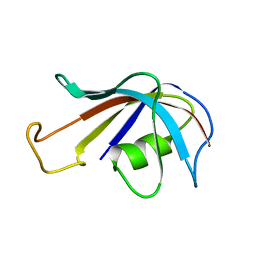 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | Descriptor: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | Authors: | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | Deposit date: | 1992-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
1FKT
 
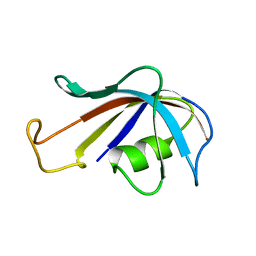 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | Descriptor: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | Authors: | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | Deposit date: | 1992-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
1FKV
 
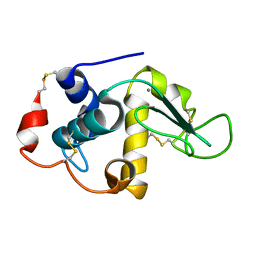 | | RECOMBINANT GOAT ALPHA-LACTALBUMIN T29I | | Descriptor: | ALPHA-LACTALBUMIN, CALCIUM ION | | Authors: | Horii, K, Matsushima, M, Tsumoto, K, Kumagai, I. | | Deposit date: | 2000-08-10 | | Release date: | 2001-02-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of Thr29 to the thermodynamic stability of goat alpha-lactalbumin as determined by experimental and theoretical approaches.
Proteins, 45, 2001
|
|
1FKW
 
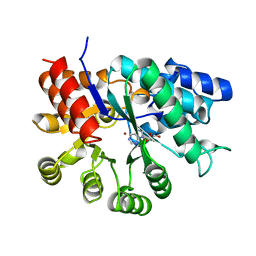 | | MURINE ADENOSINE DEAMINASE (D295E) | | Descriptor: | ADENOSINE DEAMINASE, PURINE RIBOSIDE, ZINC ION | | Authors: | Wilson, D.K, Quiocho, F.A. | | Deposit date: | 1996-02-29 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the functional role of two conserved active site aspartates in mouse adenosine deaminase.
Biochemistry, 35, 1996
|
|
1FKX
 
 | | MURINE ADENOSINE DEAMINASE (D296A) | | Descriptor: | 6-HYDROXY-1,6-DIHYDRO PURINE NUCLEOSIDE, ADENOSINE DEAMINASE, ZINC ION | | Authors: | Wilson, D.K, Quiocho, F.A. | | Deposit date: | 1996-02-29 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the functional role of two conserved active site aspartates in mouse adenosine deaminase.
Biochemistry, 35, 1996
|
|
1FKY
 
 | | NMR STUDY OF B-DNA CONTAINING A MISMATCHED BASE PAIR IN THE 29-39 K-RAS GENE SEQUENCE: CC CT C+C C+T, 2 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*TP*TP*GP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*CP*CP*AP*GP*CP*TP*C)-3') | | Authors: | Boulard, Y, Cognet, J.A.H, Fazakerley, G.V. | | Deposit date: | 1996-10-09 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure as a function of pH of two central mismatches, C . T and C . C, in the 29 to 39 K-ras gene sequence, by nuclear magnetic resonance and molecular dynamics.
J.Mol.Biol., 268, 1997
|
|
1FKZ
 
 | | NMR STUDY OF B-DNA CONTAINING A MISMATCHED BASE PAIR IN THE 29-39 K-RAS GENE SEQUENCE: CC CT C+C C+T, 2 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*TP*CP*GP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*CP*CP*AP*GP*CP*TP*C)-3') | | Authors: | Boulard, Y, Cognet, J.A.H, Fazakerley, G.V. | | Deposit date: | 1996-10-09 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure as a function of pH of two central mismatches, C . T and C . C, in the 29 to 39 K-ras gene sequence, by nuclear magnetic resonance and molecular dynamics.
J.Mol.Biol., 268, 1997
|
|
1FL0
 
 | | CRYSTAL STRUCTURE OF THE EMAP2/RNA-BINDING DOMAIN OF THE P43 PROTEIN FROM HUMAN AMINOACYL-TRNA SYNTHETASE COMPLEX | | Descriptor: | ENDOTHELIAL-MONOCYTE ACTIVATING POLYPEPTIDE II | | Authors: | Renault, L, Kerjan, P, Pasqualato, S, Menetrey, J, Robinson, J.-C, Kawaguchi, S, Vassylyev, D.G, Yokoyama, S, Mirande, M, Cherfils, J. | | Deposit date: | 2000-08-11 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the EMAPII domain of human aminoacyl-tRNA synthetase complex reveals evolutionary dimer mimicry.
EMBO J., 20, 2001
|
|
1FL1
 
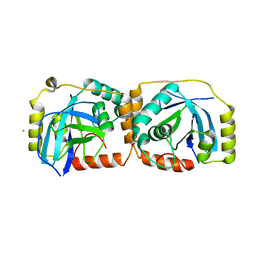 | | KSHV PROTEASE | | Descriptor: | POTASSIUM ION, PROTEASE | | Authors: | Reiling, K.K, Pray, T.R, Craik, C.S, Stroud, R.M. | | Deposit date: | 2000-08-11 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional consequences of the Kaposi's sarcoma-associated herpesvirus protease structure: regulation of activity and dimerization by conserved structural elements.
Biochemistry, 39, 2000
|
|
1FL2
 
 | |
1FL3
 
 | |
1FL5
 
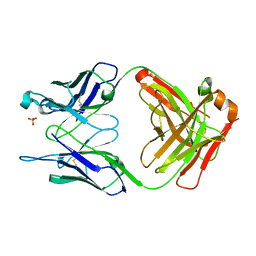 | | THE UNLIGANDED GERMLINE PRECURSOR TO THE SULFIDE OXIDASE CATALYTIC ANTIBODY 28B4. | | Descriptor: | ANTIBODY GERMLINE PRECURSOR TO ANTIBODY 28B4, SULFATE ION | | Authors: | Yin, J, Mundorff, E.C, Yang, P.L, Wendt, K.U, Hanway, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2000-08-11 | | Release date: | 2001-11-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A comparative analysis of the immunological evolution of antibody 28B4.
Biochemistry, 40, 2001
|
|
1FL6
 
 | | THE HAPTEN COMPLEXED GERMLINE PRECURSOR TO SULFIDE OXIDASE CATALYTIC ANTIBODY 28B4 | | Descriptor: | 1-[N-4'-NITROBENZYL-N-4'-CARBOXYBUTYLAMINO]METHYLPHOSPHONIC ACID, ANTIBODY GERMLINE PRECURSOR TO 28B4 | | Authors: | Yin, J, Mundorff, E.C, Yang, P.L, Wendt, K.U, Hanway, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2000-08-11 | | Release date: | 2001-11-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A comparative analysis of the immunological evolution of antibody 28B4.
Biochemistry, 40, 2001
|
|
1FL7
 
 | | HUMAN FOLLICLE STIMULATING HORMONE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLLICLE STIMULATING PROTEIN ALPHA CHAIN, ... | | Authors: | Fox, K.M, Dias, J.A, Van Roey, P. | | Deposit date: | 2000-08-11 | | Release date: | 2001-03-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structure of human follicle-stimulating hormone.
Mol.Endocrinol., 15, 2001
|
|
1FL8
 
 | |
1FL9
 
 | | THE YJEE PROTEIN | | Descriptor: | HYPOTHETICAL PROTEIN HI0065, MERCURY (II) ION | | Authors: | Teplyakov, A, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2000-08-13 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the YjeE protein from Haemophilus influenzae: a putative Atpase involved in cell wall synthesis
Proteins, 48, 2002
|
|
1FLA
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: G57D REDUCED | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1996-12-18 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
