6KMN
 
 | | Crystal Structure of Dye Decolorizing peroxidase from Bacillus subtilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Deferrochelatase/peroxidase EfeB, ... | | Authors: | Dhankhar, P, Dalal, V, Mahto, J.K, Kumar, P. | | Deposit date: | 2019-07-31 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Characterization of dye-decolorizing peroxidase from Bacillus subtilis.
Arch.Biochem.Biophys., 693, 2020
|
|
6KMM
 
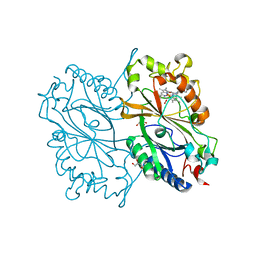 | | Crystal Structure of HEPES bound Dye Decolorizing peroxidase from Bacillus subtilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Dhankhar, P, Dalal, V, Mahto, J.K, Kumar, P. | | Deposit date: | 2019-07-31 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Characterization of dye-decolorizing peroxidase from Bacillus subtilis.
Arch.Biochem.Biophys., 693, 2020
|
|
2NCN
 
 | |
2NBW
 
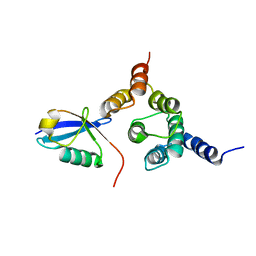 | | Solution structure of the Rpn1 T1 site with the Rad23 UBL domain | | Descriptor: | 26S proteasome regulatory subunit RPN1, UV excision repair protein RAD23 | | Authors: | Chen, X, Walters, K.J. | | Deposit date: | 2016-03-14 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome.
Structure, 24, 2016
|
|
2NB1
 
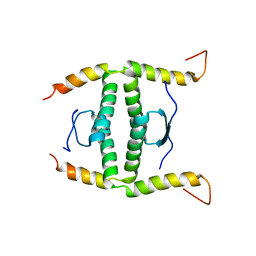 | | P63/p73 hetero-tetramerisation domain | | Descriptor: | Tumor protein 63, Tumor protein p73 | | Authors: | Gebel, J, Buchner, L, Loehr, F.M, Luh, L.M, Coutandin, D, Guentert, P, Doetsch, V. | | Deposit date: | 2016-01-19 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism of TAp73 inhibition by Delta Np63 and structural basis of p63/p73 hetero-tetramerization.
Cell Death Differ., 23, 2016
|
|
2N80
 
 | | p75NTR DD:RhoGDI | | Descriptor: | Rho GDP-dissociation inhibitor 1, Tumor necrosis factor receptor superfamily member 16 | | Authors: | Lin, Z, Ibanez, C.F. | | Deposit date: | 2015-09-30 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of death domain signaling in the p75 neurotrophin receptor
Elife, 4, 2015
|
|
2N7Y
 
 | | NMR structure of metal-binding domain 1 of ATP7B | | Descriptor: | Copper-transporting ATPase 2 | | Authors: | Yu, C, Lee, W, Dmitriev, O. | | Deposit date: | 2015-09-27 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of Metal Binding Domain 1 of the Copper Transporter ATP7B Reveals Mechanism of a Singular Wilson Disease Mutation.
Sci Rep, 8, 2018
|
|
2N5X
 
 | |
2N3T
 
 | |
2N3W
 
 | |
2N3U
 
 | |
2N3V
 
 | |
2N1R
 
 | | NMR Structure of the Myristylated Feline Immunodeficiency Virus Matrix Protein | | Descriptor: | Matrix protein p15 | | Authors: | Brown, L.A, Cox, C, Button, R.J, Baptiste, J, Bahlow, K, Spurrier, V, Luttge, B.G, Kuo, L, Freed, E.O, Summers, M.F, Kyser, J, Summers, H.R. | | Deposit date: | 2015-04-15 | | Release date: | 2015-05-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the myristylated feline immunodeficiency virus matrix protein.
Viruses, 7, 2015
|
|
2MYZ
 
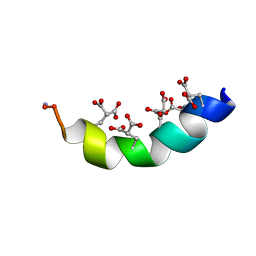 | | The Solution Structure of the Magnesium-bound Conantokin-R1B Mutant | | Descriptor: | Conantokin-R1-B | | Authors: | Kunda, S, Yuan, Y, Balsara, R.D, Zajicek, J, Castellino, F.J. | | Deposit date: | 2015-02-04 | | Release date: | 2015-06-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Hydroxyproline-induced Helical Disruption in Conantokin Rl-B Affects Subunit-selective Antagonistic Activities toward Ion Channels of N-Methyl-d-aspartate Receptors.
J.Biol.Chem., 290, 2015
|
|
2MSU
 
 | |
2ML2
 
 | |
2ML3
 
 | |
2MJC
 
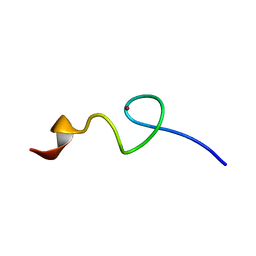 | | Zn-binding domain of eukaryotic translation initiation factor 3, subunit G | | Descriptor: | Eukaryotic translation initiation factor 3 subunit G, ZINC ION | | Authors: | Al-Abdul-Wahid, M, Menade, M, Xie, J, Kozlov, G, Gehring, K. | | Deposit date: | 2014-01-03 | | Release date: | 2015-01-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the Zn-binding domain of eukaryotic translation initiation factor 3, subunit G
To be Published
|
|
2MBC
 
 | | Solution Structure of human holo-PRL-3 in complex with vanadate | | Descriptor: | Protein tyrosine phosphatase type IVA 3 | | Authors: | Jeong, K, Kang, D, Kim, J, Shin, S, Jin, B, Lee, C, Kim, E, Jeon, Y.H, Kim, Y. | | Deposit date: | 2013-07-29 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and backbone dynamics of vanadate-bound PRL-3: comparison of 15N nuclear magnetic resonance relaxation profiles of free and vanadate-bound PRL-3.
Biochemistry, 53, 2014
|
|
2M8U
 
 | | Solution structure of the Dictyostelium discodieum Myosin Light Chain, MlcC | | Descriptor: | Myosin Light Chain, MlcC | | Authors: | Liburd, J.D, Miller, E, Langelaan, D, Chitayat, S, Crawley, S.W, Cote, G.P, Smith, S.P. | | Deposit date: | 2013-05-28 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Single-lobe Myosin Light Chain C in Complex with the Light Chain-binding Domains of Myosin-1C Provides Insights into Divergent IQ Motif Recognition.
J.Biol.Chem., 291, 2016
|
|
2M8I
 
 | | Structure of Pin1 WW domain | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Luh, L.M, Kirchner, D.K, Loehr, F, Haensel, R, Doetsch, V. | | Deposit date: | 2013-05-22 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular crowding drives active Pin1 into nonspecific complexes with endogenous proteins prior to substrate recognition.
J.Am.Chem.Soc., 135, 2013
|
|
2M8J
 
 | | Structure of Pin1 WW domain phospho-mimic S16E | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Luh, L.M, Kirchner, D.K, Loehr, F, Haensel, R, Doetsch, V. | | Deposit date: | 2013-05-22 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular crowding drives active Pin1 into nonspecific complexes with endogenous proteins prior to substrate recognition.
J.Am.Chem.Soc., 135, 2013
|
|
2M85
 
 | | PHD Domain from Human SHPRH | | Descriptor: | E3 ubiquitin-protein ligase SHPRH, ZINC ION | | Authors: | Machado, L.E.S.F, Pustovalova, Y, Pozhidaeva, A, Almeida, F.C.L, Bezsonova, I, Korzhnev, D.M. | | Deposit date: | 2013-05-07 | | Release date: | 2013-08-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | PHD domain from human SHPRH.
J.Biomol.Nmr, 56, 2013
|
|
2M4H
 
 | | Solution structure of the Core Domain (10-76) of the Feline Calicivirus VPg protein | | Descriptor: | Feline Calicivirus VPg protein | | Authors: | Kwok, R.N, Leen, E.N, Birtley, J.R, Prater, S.N, Simpson, P.J, Curry, S, Matthews, S, Marchant, J. | | Deposit date: | 2013-02-05 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of the Compact Helical Core Domains of Feline Calicivirus and Murine Norovirus VPg Proteins.
J.Virol., 87, 2013
|
|
2LX9
 
 | |
