9IUZ
 
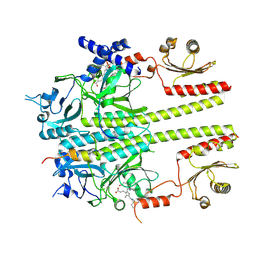 | | Constitutively active mutant(Y276H) of Arabidopsis phytochrome B(phyB) in complex with phytochrome-interacting factor 6(PIF6) | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome B, Phytochrome-interacting factor 6 | | Authors: | Wang, Z, Wang, W, Zhao, D, Song, Y, Xu, B, Zhao, J, Wang, J. | | Deposit date: | 2024-07-22 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor.
Cell, 2024
|
|
9EUY
 
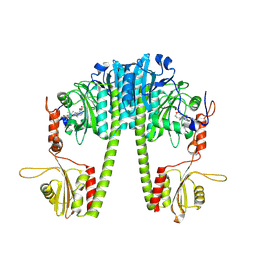 | | Cryo-EM structure of the full-length Pseudomonas aeruginosa bacteriophytochrome in its Pfr state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Bodizs, S, Westenhoff, S. | | Deposit date: | 2024-03-28 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-EM structures of a bathy phytochrome histidine kinase reveal a unique light-dependent activation mechanism.
Structure, 2024
|
|
9EUT
 
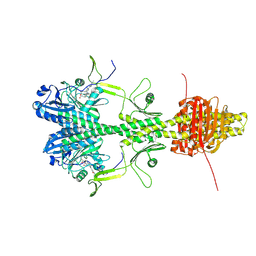 | | Cryo-EM structure of the full-length Pseudomonas aeruginosa bacteriophytochrome in its Pr state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Bodizs, S, Westenhoff, S. | | Deposit date: | 2024-03-28 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-EM structures of a bathy phytochrome histidine kinase reveal a unique light-dependent activation mechanism.
Structure, 2024
|
|
8W26
 
 | |
8YB4
 
 | | Pfr conformer of Arabidopsis thaliana phytochrome B in complex with phytochrome-interacting factor 6 | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, phytochrome B, phytochrome-interacting factor 6 | | Authors: | Wang, Z, Wang, W, Zhao, D, Song, Y, Xu, B, Zhao, J, Wang, J. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor.
Cell, 2024
|
|
8UPM
 
 | |
8UQK
 
 | |
8UQI
 
 | |
8UPK
 
 | |
8UPH
 
 | |
8ULG
 
 | | Cryo-EM structure of bovine phosphodiesterase 6 bound to IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
8UGS
 
 | | Cryo-EM structure of bovine phosphodiesterase 6 bound to cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
8UGB
 
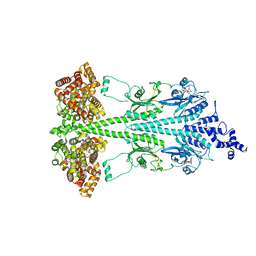 | | Cryo-EM structure of bovine phosphodiesterase 6 bound to udenafil | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-05 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
8UFI
 
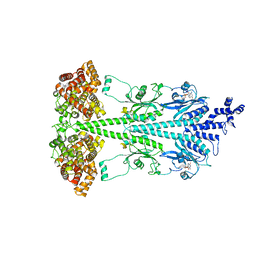 | | Cryo-EM structure of bovine phosphodiesterase 6 | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-04 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
8U8Z
 
 | | Cryo-EM structure of PsBphP in Pr state, extended DHp | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-18 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
8U65
 
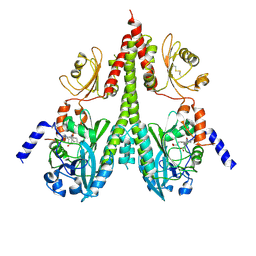 | | Cryo-EM structure of PsBphP in Pfr state, splayed PSM only | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-13 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
8U64
 
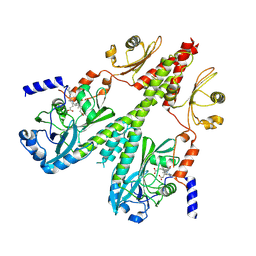 | | Cryo-EM structure of PsBphP in Pfr state, medial PSM only | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-13 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
8U63
 
 | | Cryo-EM structure of PsBphP in Pfr state, Dimer of Dimers PSM only | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-13 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
8U62
 
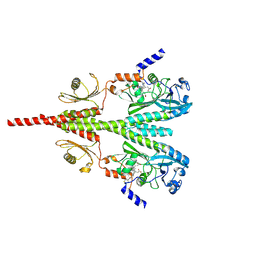 | | Cryo-EM structure of PsBphP in Pfr state, Dimer of Dimers FL | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-13 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
8U4X
 
 | | Cryo-EM structure of PsBphP in Pr state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-11 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
8K9O
 
 | |
8JRC
 
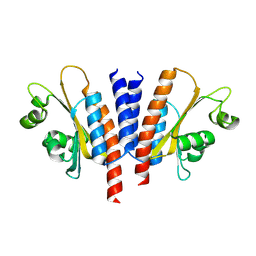 | |
8SGK
 
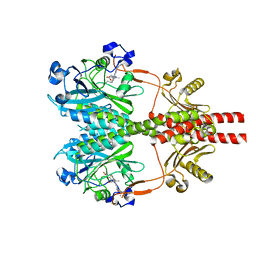 | | CryoEM structure of Deinococcus radiodurans BphP photosensory module in Pr state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Li, H, Li, H. | | Deposit date: | 2023-04-12 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | CryoEM structure of Deinococcus radiodurans BphP photosensory module in Pr state
To Be Published
|
|
8ISK
 
 | | Pr conformer of Zea mays phytochrome A1 - ZmphyA1-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
8ISJ
 
 | | Pr conformer of Arabidopsis thaliana phytochrome A - AtphyA-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
