3D7O
 
 | | Human hemoglobin, nitrogen dioxide anion modified | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, NITRITE ION, ... | | Authors: | Yi, J, Safo, M.K, Richter-Addo, G.B. | | Deposit date: | 2008-05-21 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The nitrite anion binds to human hemoglobin via the uncommon O-nitrito mode.
Biochemistry, 47, 2008
|
|
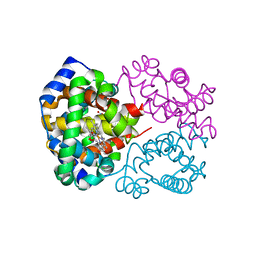
2UVG
 
 | |
2VFZ
 
 | | CRYSTAL STRUCTURE OF ALPHA-1,3 GALACTOSYLTRANSFERASE (R365K) IN COMPLEX WITH UDP-2F-GALACTOSE | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYL TRANSFERASE, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUOROGALACTOSE | | Authors: | Jamaluddin, H, Tumbale, P, Withers, S.G, Acharya, K.R, Brew, K. | | Deposit date: | 2007-11-06 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Changes Induced by Binding Udp-2F-Galactose to Alpha-1,3 Galactosyltransferase-Implications for Catalysis
J.Mol.Biol., 369, 2007
|
|
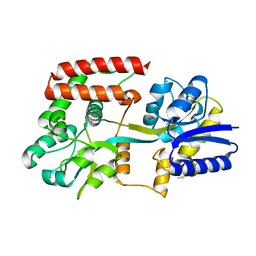
2O69
 
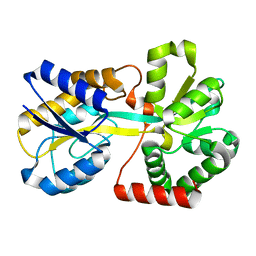 | |
2OJW
 
 | | Crystal structure of human glutamine synthetase in complex with ADP and phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Karlberg, T, Uppenberg, J, Arrowsmith, C, Berglund, H, Busam, R.D, Collins, R, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Hogbom, M, Johansson, I, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Wallden, K, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-15 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design.
J.Mol.Biol., 375, 2008
|
|
3O1D
 
 | | Structure-function study of Gemini derivatives with two different side chains at C-20, Gemini-0072 and Gemini-0097. | | Descriptor: | (1R,3R,7E,17beta)-17-[(1S)-6,6,6-trifluoro-5-hydroxy-1-(4-hydroxy-4-methylpentyl)-5-(trifluoromethyl)hex-3-yn-1-yl]-9,10-secoestra-5,7-diene-1,3-diol, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | Authors: | Huet, T, Moras, D, Rochel, N. | | Deposit date: | 2010-07-21 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4007 Å) | | Cite: | Structure-function study of gemini derivatives with two different side chains at C-20, Gemini-0072 and Gemini-0097.
Medchemcomm, 2, 2011
|
|
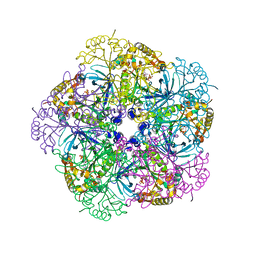
3KBM
 
 | |
2ZCE
 
 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with pyrrolysine and an ATP analogue | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PYRROLYSINE, ... | | Authors: | Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-11-08 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Studies on Multiple Conformational States of Active-site Loops in Pyrrolysyl-tRNA Synthetase
J.Mol.Biol., 378, 2008
|
|
7UGF
 
 | | First bromodomain of BRD4 liganded with BMS-536924 | | Descriptor: | (3M)-4-{[(2S)-2-(3-chlorophenyl)-2-hydroxyethyl]amino}-3-[4-methyl-6-(morpholin-4-yl)-1H-benzimidazol-2-yl]pyridin-2(1H)-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2022-03-24 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis of CBP and EP300 interaction with kinase inhibitors
To Be Published
|
|
7UG5
 
 | | Second bromodomain of BRD3 liganded with BMS-536924 | | Descriptor: | (3M)-4-{[(2S)-2-(3-chlorophenyl)-2-hydroxyethyl]amino}-3-[4-methyl-6-(morpholin-4-yl)-1H-benzimidazol-2-yl]pyridin-2(1H)-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 3 | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2022-03-23 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of CBP and EP300 interaction with kinase inhibitors
To Be Published
|
|
3KCL
 
 | |
7UGL
 
 | | Bromodomain of CBP liganded with BMS-536924 and SGC-CBP30 | | Descriptor: | (3M)-4-{[(2S)-2-(3-chlorophenyl)-2-hydroxyethyl]amino}-3-[4-methyl-6-(morpholin-4-yl)-1H-benzimidazol-2-yl]pyridin-2(1H)-one, 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, Histone acetyltransferase | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2022-03-24 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of CBP and EP300 interaction with kinase inhibitors
To Be Published
|
|
6U72
 
 | | BRD4-BD1 in complex with the cyclic peptide 3.1_2_AcK5toA | | Descriptor: | 3.1_2_AcK5toA, AMINO GROUP, Bromodomain-containing protein 4 | | Authors: | Patel, K, Walshe, J.L, Walport, L.J, Mackay, J.P, Mouradian, K.S. | | Deposit date: | 2019-08-31 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cyclic peptides can engage a single binding pocket through highly divergent modes.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2P6W
 
 | | Crystal structure of a glycosyltransferase involved in the glycosylation of the major capsid of PBCV-1 | | Descriptor: | CITRATE ANION, MANGANESE (II) ION, Putative glycosyltransferase (Mannosyltransferase) involved in glycosylating the PBCV-1 major capsid protein | | Authors: | Zhang, Y, Ye, X, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2007-03-19 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and function of a chlorella virus-encoded glycosyltransferase.
Structure, 15, 2007
|
|
7S10
 
 | | Crystal Structure of ascorbate peroxidase triple mutant: S160M, L203M, Q204M | | Descriptor: | L-ascorbate peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Poulos, T.L, Kim, J, Murarka, V.C. | | Deposit date: | 2021-08-31 | | Release date: | 2022-09-07 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.40000689 Å) | | Cite: | Computational analysis of the tryptophan cation radical energetics in peroxidase Compound I.
J.Biol.Inorg.Chem., 27, 2022
|
|
6EM0
 
 | | Crystal Structure of 2-hydroxybiphenyl 3-monooxygenase M321A from Pseudomonas azelaica | | Descriptor: | 2-hydroxybiphenyl-3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Deri, B, Bregman-Cohen, A, Pazy Benhar, Y, Fishman, A. | | Deposit date: | 2017-10-01 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Altering 2-Hydroxybiphenyl 3-Monooxygenase Regioselectivity by Protein Engineering for the Production of a New Antioxidant.
Chembiochem, 19, 2018
|
|
7VB6
 
 | | Crystal structure of hydroxynitrile lyase from Linum usitatissium complexed with (R)-2-hydroxy-2-methylbutanenitrile | | Descriptor: | (2R)-2-methyl-2-oxidanyl-butanenitrile, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zheng, D, Nakabayashi, M, Asano, Y. | | Deposit date: | 2021-08-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural characterization of Linum usitatissimum hydroxynitrile lyase: A new cyanohydrin decomposition mechanism involving a cyano-zinc complex.
J.Biol.Chem., 298, 2022
|
|
6D03
 
 | | Cryo-EM structure of a Plasmodium vivax invasion complex essential for entry into human reticulocytes; one molecule of parasite ligand. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(2-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruszczyk, J, Huang, R.K, Hong, C, Yu, Z, Tham, W.H. | | Deposit date: | 2018-04-10 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
3KBN
 
 | |
6EYD
 
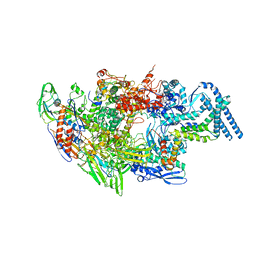 | | Structure of Mycobacterium smegmatis RNA polymerase Sigma-A holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Kouba, T, Barvik, I, Krasny, L. | | Deposit date: | 2017-11-11 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | The Core and Holoenzyme Forms of RNA Polymerase fromMycobacterium smegmatis.
J. Bacteriol., 201, 2019
|
|
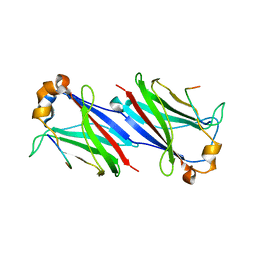
6F8G
 
 | |
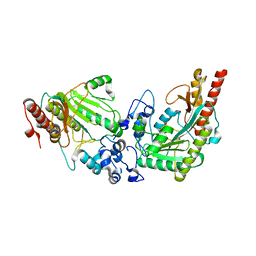
3L2D
 
 | |
4LWU
 
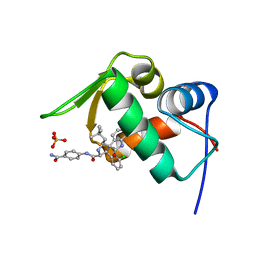 | | The 1.14A Crystal Structure of Humanized Xenopus MDM2 with RO5499252 | | Descriptor: | (2'S,3R,4'S,5'R)-N-(4-carbamoylphenyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Graves, B.J, Lukacs, C, Janson, C.A. | | Deposit date: | 2013-07-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Discovery of potent and selective spiroindolinone MDM2 inhibitor, RO8994, for cancer therapy.
Bioorg.Med.Chem., 22, 2014
|
|
7L2N
 
 | |
7L2O
 
 | |
