7FZT
 
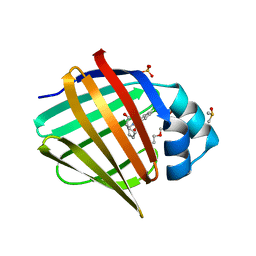 | | Crystal Structure of human FABP4 binding site mutated to that of FABP5 in complex with rac-(1R,2S)-2-[(3,4-dichlorobenzoyl)amino]cyclohexane-1-carboxylic acid, i.e. SMILES C1CC[C@@H]([C@@H](C1)C(=O)O)NC(=O)c1cc(c(cc1)Cl)Cl with IC50=15.8182 microM | | Descriptor: | (1R,2S)-2-(3,4-dichlorobenzamido)cyclohexane-1-carboxylic acid, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Buettelmann, B, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
9GPY
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12g/j | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclohexyl-18,19-dimethoxy-13-methyl-11,17-dioxa-4-aza-1lambda5,18lambda5-diphospha-1,15-distannatetracyclo[13.4.0.04,9.016,20]nonadecane-3,10-quinone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GQ2
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog p5(1,1)-(E) | | Descriptor: | (2~{S},9~{S},13~{E})-2-cyclohexyl-18,21-dimethoxy-11,16-dioxa-4-azatricyclo[15.2.2.0^{4,9}]henicosa-1(19),13,17,20-tetraene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
7PUG
 
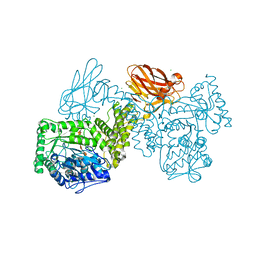 | | GH115 alpha-1,2-glucuronidase in complex with xylopentaose | | Descriptor: | CALCIUM ION, CHLORIDE ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Wilkens, C, Morth, J.P, Polikarpov, I. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | A GH115 alpha-glucuronidase structure reveals dimerization-mediated substrate binding and a proton wire potentially important for catalysis.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
3JVL
 
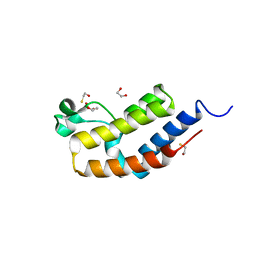 | | Crystal structure of bromodomain 2 of mouse Brd4 | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Bromodomain-containing protein 4 | | Authors: | Vollmuth, F, Blankenfeldt, W, Geyer, M. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structures of the Dual Bromodomains of the P-TEFb-activating Protein Brd4 at Atomic Resolution
J.Biol.Chem., 284, 2009
|
|
9N3R
 
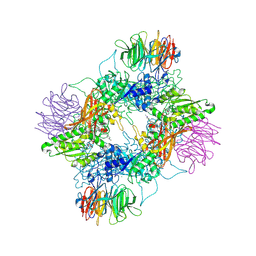 | | Crystal structure of PRMT5:MEP50 in complex with MTA and TNG462 | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, CHLORIDE ION, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2025-01-31 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Discovery of TNG462: A Highly Potent and Selective MTA-Cooperative PRMT5 Inhibitor to Target Cancers with MTAP Deletion.
J.Med.Chem., 68, 2025
|
|
8H9F
 
 | | Human ATP synthase state 1 subregion 3 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ATP synthase F(0) complex subunit B1, mitochondrial, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-08-30 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
7QA4
 
 | |
6RBO
 
 | |
6RC5
 
 | |
6C3B
 
 | | O2-, PLP-Dependent L-Arginine Hydroxylase RohP Holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, TRIETHYLENE GLYCOL, Uncharacterized protein | | Authors: | Hedges, J.B, Ryan, K.S. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Snapshots of the Catalytic Cycle of an O2, Pyridoxal Phosphate-Dependent Hydroxylase.
ACS Chem. Biol., 13, 2018
|
|
9N3P
 
 | |
8AOM
 
 | | Complex of PD-L1 with VHH1 | | Descriptor: | MAGNESIUM ION, Programmed cell death 1 ligand 1, VHH6 | | Authors: | Kang-Pettinger, T, Hall, G. | | Deposit date: | 2022-08-08 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Identification, binding, and structural characterization of single domain anti-PD-L1 antibodies inhibitory of immune regulatory proteins PD-1 and CD80.
J.Biol.Chem., 299, 2023
|
|
5GV1
 
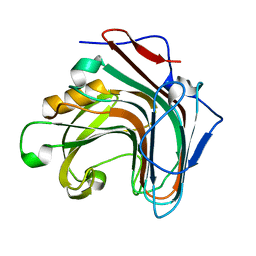 | | Crystal structure of ENZbleach xylanase wild type | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Chitnumsub, P, Jaruwat, A, Boonyapakorn, K, Noytanom, K. | | Deposit date: | 2016-09-01 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based protein engineering for thermostable and alkaliphilic enhancement of endo-beta-1,4-xylanase for applications in pulp bleaching
J. Biotechnol., 259, 2017
|
|
6ZTQ
 
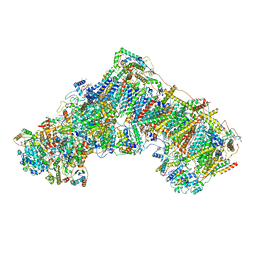 | | Cryo-EM structure of respiratory complex I from Mus musculus inhibited by piericidin A at 3.0 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bridges, H.R, Blaza, J.N, Agip, A.N.A, Hirst, J. | | Deposit date: | 2020-07-20 | | Release date: | 2020-10-21 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of inhibitor-bound mammalian complex I.
Nat Commun, 11, 2020
|
|
4AAF
 
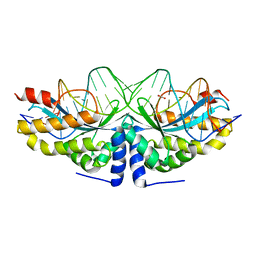 | | Crystal structure of the mutant D75N I-CreI in complex with an altered target (The four central bases, 2NN region, are composed by TGCA from 5' to 3') | | Descriptor: | 1,2-ETHANEDIOL, 24MER DNA, DNA ENDONUCLEASE I-CREI | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F.L, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
6H78
 
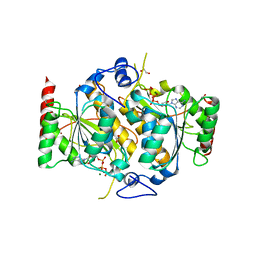 | | E1 enzyme for ubiquitin like protein activation. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Soudah, N, Padala, P, Hassouna, F, Mashahreh, B, Lebedev, A.A, Isupov, M.N, Cohen-Kfir, E, Wiener, R. | | Deposit date: | 2018-07-30 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An N-Terminal Extension to UBA5 Adenylation Domain Boosts UFM1 Activation: Isoform-Specific Differences in Ubiquitin-like Protein Activation.
J.Mol.Biol., 431, 2019
|
|
8JAM
 
 | |
6OSY
 
 | |
9MGL
 
 | |
5GZP
 
 | |
8AQ4
 
 | | In surfo structure of the membrane integral lipoprotein N-acyltransferase Lnt from E. coli in complex with TITC and lyso-PE | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Huang, C.-Y, Weichert, D, Boland, C, Smithers, L, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8VTV
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with macrolone MCX-91, mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.55A resolution | | Descriptor: | 1-cyclopropyl-7-{7-[({[(3aR,4R,7R,8S,9S,10R,11R,13R,14E,15S,15aR)-10-{[(2S,3R,4S,6R)-4-(dimethylamino)-3-hydroxy-6-methyloxan-2-yl]oxy}-4-ethyl-11-methoxy-14-(methoxyimino)-3a,7,9,11,13,15-hexamethyl-2,6-dioxododecahydro-2H,4H-[1,3]dioxolo[4,5-c]oxacyclotetradecin-8-yl]oxy}carbonyl)amino]hept-1-yn-1-yl}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Ma, C.-X, Klepacki, D, Alizadeh, F, Vazquez-Laslop, N, Liang, J.-H, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2024-01-27 | | Release date: | 2024-08-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms.
Nat.Chem.Biol., 20, 2024
|
|
6I5H
 
 | | Crystal structure of CLK1 in complex with furanopyrimidin VN412 | | Descriptor: | 1,2-ETHANEDIOL, 5-(1-methylpyrazol-4-yl)-3-(3-phenoxyphenyl)furo[3,2-b]pyridine, Dual specificity protein kinase CLK1, ... | | Authors: | Schroeder, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6I5Q
 
 | | Papaver somniferum O-methyltransferase 1 | | Descriptor: | O-methyltransferase 1 | | Authors: | Cabry, M.P, Offen, W.A, Winzer, T, Li, Y, Graham, I.A, Davies, G.J, Saleh, P. | | Deposit date: | 2018-11-14 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of Papaver somniferum O-Methyltransferase 1 Reveals Initiation of Noscapine Biosynthesis with Implications for Plant Natural Product Methylation
Acs Catalysis, 2019
|
|
