2ZEY
 
 | | Family 16 carbohydrate binding module | | Descriptor: | CALCIUM ION, S-layer associated multidomain endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose, ... | | Authors: | Nair, S.K, Bae, B. | | Deposit date: | 2007-12-18 | | Release date: | 2008-03-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Basis for the Selectivity and Specificity of Ligand Recognition by the Family 16 Carbohydrate-binding Modules from Thermoanaerobacterium polysaccharolyticum ManA
J.Biol.Chem., 283, 2008
|
|
7QWV
 
 | | Crystal structure of the REC114-TOPOVIBL complex. | | Descriptor: | Meiotic recombination protein REC114, Type 2 DNA topoisomerase 6 subunit B-like | | Authors: | Juarez-Martinez, A.B, Robert, T, de Massy, B, Kadlec, J. | | Deposit date: | 2022-01-25 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | TOPOVIBL-REC114 interaction regulates meiotic DNA double-strand breaks.
Nat Commun, 13, 2022
|
|
3T81
 
 | |
2ZAW
 
 | | Crystal Structure of Ferric Cytochrome P450cam Reconstituted with 6-Methyl-6-depropionated Hemin | | Descriptor: | 6-METHY-6-DEPROPIONATEHEMIN, CAMPHOR, CHLORIDE ION, ... | | Authors: | Harada, K, Sakurai, K, Shimada, H, Tsukihara, T, Hayashi, T. | | Deposit date: | 2007-10-11 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Evaluation of the functional role of the heme-6-propionate side chain in cytochrome P450cam
J.Am.Chem.Soc., 130, 2008
|
|
2ZJ5
 
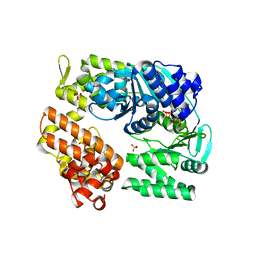 | | Archaeal DNA helicase Hjm complexed with ADP in form 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Putative ski2-type helicase, SULFATE ION | | Authors: | Oyama, T, Oka, H, Fujikane, R, Ishino, Y, Morikawa, K. | | Deposit date: | 2008-02-29 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic structures and functional implications of the archaeal RecQ-like helicase Hjm
Bmc Struct.Biol., 9, 2009
|
|
1MZW
 
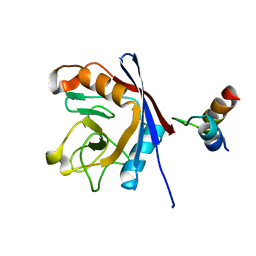 | | Crystal structure of a U4/U6 snRNP complex between human spliceosomal cyclophilin H and a U4/U6-60K peptide | | Descriptor: | U-snRNP-associated cyclophilin, U4/U6 snrnp 60kDa protein | | Authors: | Reidt, U, Wahl, M.C, Horowitz, D.S, Luehrmann, R, Ficner, R. | | Deposit date: | 2002-10-10 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a complex between human spliceosomal cyclophilin H and a U4/U6 snRNP-60K peptide
J.Mol.Biol., 331, 2003
|
|
2ZE0
 
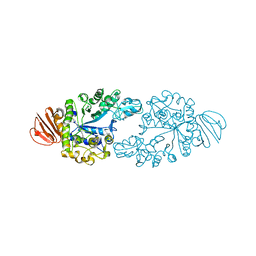 | | Alpha-glucosidase GSJ | | Descriptor: | Alpha-glucosidase, CALCIUM ION | | Authors: | Shirai, T, Hung, V.S, Morinaka, K, Kobayashi, T, Ito, S. | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GH13 alpha-glucosidase GSJ from one of the deepest sea bacteria
Proteins, 73, 2008
|
|
3T9J
 
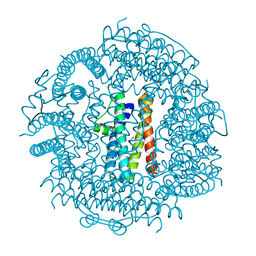 | | Crystal structure HP-NAP from strain YS39 in apo form | | Descriptor: | 1,2-ETHANEDIOL, Neutrophil-activating protein | | Authors: | Tsuruta, O, Yokoyama, H, Fujii, S. | | Deposit date: | 2011-08-03 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A new crystal lattice structure of Helicobacter pylori neutrophil-activating protein (HP-NAP)
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3TA9
 
 | | beta-Glucosidase A from the halothermophile H. orenii | | Descriptor: | Glycoside hydrolase family 1, NICKEL (II) ION | | Authors: | Hofmann, A, Wang, C.K, Kori, L.D, Patel, B.K. | | Deposit date: | 2011-08-03 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Expression, purification and preliminary crystallographic analysis of the recombinant (beta)-glucosidase (BglA) from the halothermophile Halothermothrix orenii
Acta Crystallogr.,Sect.F, 67, 2011
|
|
6BTY
 
 | | Crystal structure of the PI3KC2alpha C2 domain in space group P41212 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha | | Authors: | Chen, K.-E, Collins, B.M. | | Deposit date: | 2017-12-08 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.678 Å) | | Cite: | Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2 alpha.
Structure, 26, 2018
|
|
2ZID
 
 | | Crystal structure of dextran glucosidase E236Q complex with isomaltotriose | | Descriptor: | CALCIUM ION, Dextran glucosidase, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose | | Authors: | Hondoh, H, Saburi, W, Mori, H, Okuyama, M, Nakada, T, Matsuura, Y, Kimura, A. | | Deposit date: | 2008-02-14 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate recognition mechanism of alpha-1,6-glucosidic linkage hydrolyzing enzyme, dextran glucosidase from Streptococcus mutans.
J.Mol.Biol., 378, 2008
|
|
1N0Y
 
 | | Crystal Structure of Pb-bound Calmodulin | | Descriptor: | ACETATE ION, CACODYLATE ION, Calmodulin, ... | | Authors: | Wilson, M.A, Brunger, A.T. | | Deposit date: | 2002-10-15 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Domain flexibility in the 1.75 A resolution structure of Pb2+-calmodulin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3TG0
 
 | | E. coli alkaline phosphatase with bound inorganic phosphate | | Descriptor: | Alkaline phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Bobyr, E, Lassila, J.K, Wiersma-Koch, H.I, Fenn, T.D, Lee, J.J, Nikolic-Hughes, I, Hodgson, K.O, Rees, D.C, Hedman, B, Herschlag, D. | | Deposit date: | 2011-08-16 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution analysis of Zn(2+) coordination in the alkaline phosphatase superfamily by EXAFS and x-ray crystallography.
J.Mol.Biol., 415, 2012
|
|
3THH
 
 | |
2ZNW
 
 | | Crystal Structure of ScFv10 in Complex with Hen Egg Lysozyme | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Lysozyme C, ScFv10 | | Authors: | DeSantis, M.E, Acchione, M, Li, M, Walter, R.L, Wlodawer, A, Smith-Gill, S. | | Deposit date: | 2008-05-02 | | Release date: | 2009-01-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Specific fluorine labeling of the HyHEL10 antibody affects antigen binding and dynamics
Biochemistry, 51, 2012
|
|
3THW
 
 | | Human MutSbeta complexed with an IDL of 4 bases (Loop4) and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA Loop4 hairpin, DNA mismatch repair protein Msh2, ... | | Authors: | Yang, W. | | Deposit date: | 2011-08-19 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Mechanism of mismatch recognition revealed by human MutSbeta bound to unpaired DNA loops
Nat.Struct.Mol.Biol., 19, 2012
|
|
3STG
 
 | | Crystal structure of A58P, DEL(N59), and loop 7 truncated mutant of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION | | Authors: | Allison, T.M, Jameson, G.B, Parker, E.J. | | Deposit date: | 2011-07-09 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An Extended (beta)7(alpha)7 Substrate-Binding Loop Is Essential for Efficient Catalysis by 3-Deoxy-D-manno-Octulosonate 8-Phosphate Synthase
Biochemistry, 50, 2011
|
|
1MRF
 
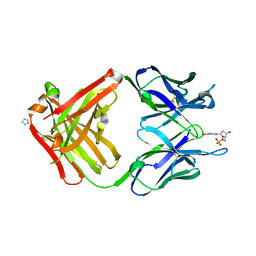 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | Descriptor: | 2'-DEOXYINOSINE-5'-MONOPHOSPHATE, IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), ... | | Authors: | Pokkuluri, P.R, Cygler, M. | | Deposit date: | 1994-06-13 | | Release date: | 1995-02-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
2ZR7
 
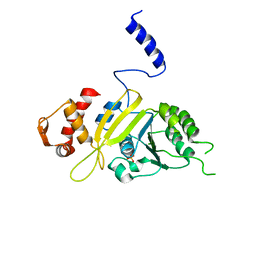 | | Msreca native form II' | | Descriptor: | PHOSPHATE ION, Protein recA | | Authors: | Prabu, J.R, Manjunath, G.P, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2008-08-25 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Functionally important movements in RecA molecules and filaments: studies involving mutation and environmental changes
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2ZRF
 
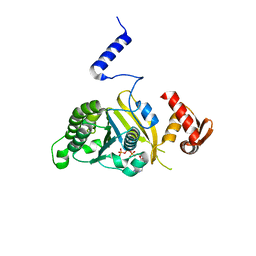 | | MsRecA Q196N dATP form IV | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Protein recA | | Authors: | Prabu, J.R, Manjunath, G.P, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2008-08-27 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functionally important movements in RecA molecules and filaments: studies involving mutation and environmental changes
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2ZRN
 
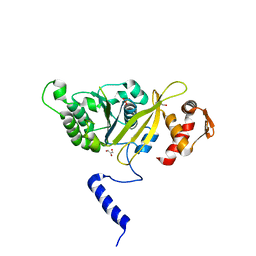 | | MsRecA Form IV | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein recA | | Authors: | Prabu, J.R, Manjunath, G.P, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2008-08-27 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Functionally important movements in RecA molecules and filaments: studies involving mutation and environmental changes
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3STE
 
 | | Crystal structure of a mutant (Q202A) of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION, SODIUM ION | | Authors: | Allison, T.M, Jameson, G.B, Gloyne, B.J, Parker, E.J. | | Deposit date: | 2011-07-09 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An Extended (beta)7(alpha)7 Substrate-Binding Loop Is Essential for Efficient Catalysis by 3-Deoxy-D-manno-Octulosonate 8-Phosphate Synthase
Biochemistry, 50, 2011
|
|
6C0U
 
 | | Crystal structure of cAMP-dependent protein kinase Calpha subunit bound with N46 | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3R,4R)-4-{[4-(2-fluoro-3-methoxy-6-propoxybenzene-1-carbonyl)benzene-1-carbonyl]amino}pyrrolidin-3-yl]-1H-indazole-5-carboxamide, PHOSPHATE ION, ... | | Authors: | Qin, L, Sankaran, B, Kim, C. | | Deposit date: | 2018-01-02 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for selective inhibition of human PKG I alpha by the balanol-like compound N46.
J. Biol. Chem., 293, 2018
|
|
3SUL
 
 | | Crystal structure of cerato-platanin 3 from M. perniciosa (MpCP3) | | Descriptor: | Cerato-platanin-like protein | | Authors: | Oliveira, J.F, Barsottini, M.R.O, Zaparoli, G, Machado, L.O, Dias, S.M.G, Pereira, G.A.G, Ambrosio, A.L.B. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization.
Mol. Plant Microbe Interact., 26, 2013
|
|
3B9X
 
 | |
