3IDJ
 
 | |
5KO5
 
 | | Crystal Structure of Isoform 2 of Purine Nucleoside Phosphorylase from Schistosoma mansoni in complex with cytosine | | Descriptor: | 1,2-ETHANEDIOL, 6-AMINOPYRIMIDIN-2(1H)-ONE, Purine nucleoside phosphorylase | | Authors: | Torini, J.R, Romanello, L, Bird, L, Owens, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The molecular structure of Schistosoma mansoni PNP isoform 2 provides insights into the nucleoside selectivity of PNPs.
PLoS ONE, 13, 2018
|
|
8P2F
 
 | | Staphylococcus aureus 70S ribosome with elongation factor G locked with fusidic acid cyclopentane in post-translocational state | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA, 2-[(3~{R},4~{S},5~{S},8~{S},9~{S},10~{S},11~{R},13~{S},14~{S},16~{S})-16-acetyloxy-4,8,10,14-tetramethyl-3,11-bis(oxidanyl)-1,2,3,4,5,6,7,9,11,12,13,15,16,17-tetradecahydrocyclopenta[a]phenanthren-17-yl]-5-cyclopentyl-pentanoic acid, ... | | Authors: | Gonzalez-Lopez, A, Selmer, M. | | Deposit date: | 2023-05-16 | | Release date: | 2024-05-01 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Structures of the Staphylococcus aureus ribosome inhibited by fusidic acid and fusidic acid cyclopentane.
Sci Rep, 14, 2024
|
|
7RU6
 
 | |
7BMO
 
 | | HEWL in cesium chloride (0.25 M CsCl in protein buffer) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Hen egg white lysozyme, ... | | Authors: | Koelmel, W, Kuper, J, Kisker, C. | | Deposit date: | 2021-01-20 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cesium based phasing of macromolecules: a general easy to use approach for solving the phase problem.
Sci Rep, 11, 2021
|
|
8DS3
 
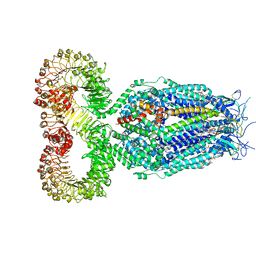 | | LRRC8A:C conformation 1 (round) | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8B5A
 
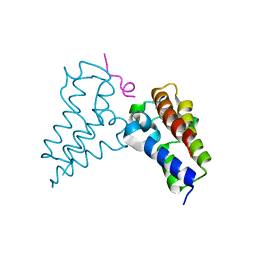 | | Human BRD3 bromodomain 2 in complex with a H4 peptide containing ApmTri (H4K20ApmTri) | | Descriptor: | Bromodomain-containing protein 3, H4K20ApmTri | | Authors: | Braun, M.B, Bartlick, N, Stehle, T. | | Deposit date: | 2022-09-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Synthesis, Biochemical Characterization, and Genetic Encoding of a 1,2,4-Triazole Amino Acid as an Acetyllysine Mimic for Bromodomains of the BET Family.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5G1W
 
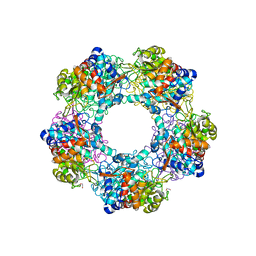 | | Apo Structure of Linalool Dehydratase-Isomerase | | Descriptor: | 1,2-ETHANEDIOL, LINALOOL DEHYDRATASE/ISOMERASE, METHYLMALONIC ACID | | Authors: | Chambers, S, Hau, A, Man, H, Omar, M, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-03-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and functional insights into asymmetric enzymatic dehydration of alkenols.
Nat. Chem. Biol., 13, 2017
|
|
7V91
 
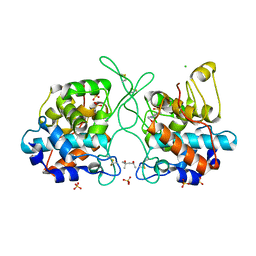 | | Crystal Structure of the Catalytic Domain of a Family GH19 Chitinase from Gazyumaru, Ficus microcarpa | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Kozome, D, Kubota, T, Ishikawa, K. | | Deposit date: | 2021-08-24 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Analysis and Construction of a Thermostable Antifungal Chitinase.
Appl.Environ.Microbiol., 88, 2022
|
|
1Q9A
 
 | | Crystal structure of the sarcin/ricin domain from E.coli 23S rRNA at 1.04 resolution | | Descriptor: | Sarcin/ricin 23S rRNA | | Authors: | Correll, C.C, Beneken, J, Plantinga, M.J, Lubbers, M, Chan, Y.L. | | Deposit date: | 2003-08-22 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | The common and distinctive features of the bulged-G motif based on a 1.04 A resolution RNA structure
Nucleic Acids Res., 31, 2003
|
|
6UII
 
 | |
5UAI
 
 | |
8FHS
 
 | | Human L-type voltage-gated calcium channel Cav1.2 in the presence of amiodarone and sofosbuvir at 3.3 Angstrom resolution | | Descriptor: | (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-13 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
6KTL
 
 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, complexed with NAD and myo-inositol, from Paracoccus laeviglucosivorans | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, ACETATE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
5G3P
 
 | | Bacillus cereus formamidase (BceAmiF) acetylated at the active site. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gavira, J.A, Conejero-Muriel, M, Martinez-Rodriguez, S. | | Deposit date: | 2016-04-29 | | Release date: | 2017-04-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A novel cysteine carbamoyl-switch is responsible for the inhibition of formamidase, a nitrilase superfamily member.
Arch.Biochem.Biophys., 662, 2019
|
|
8AK2
 
 | | Drosophila melanogaster UNC89 Protein Kinase Domain 1 (apo) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Obscurin | | Authors: | Dorendorf, T, Zacharchenko, T, Mayans, O. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PK1 from Drosophila obscurin is an inactive pseudokinase with scaffolding properties.
Open Biology, 13, 2023
|
|
7MOV
 
 | | PTP1B 1-301 F225Y-R199N mutations | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-05-03 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
4WK4
 
 | | Metal Ion and Ligand Binding of Integrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALA-CYS-ARG-GLY-ASP-GLY-TRP-CYS, ... | | Authors: | Xia, W, Springer, T.A. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Metal ion and ligand binding of integrin alpha 5 beta 1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5ED4
 
 | | Structure of a PhoP-DNA complex | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Wang, S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of DNA sequence recognition by the response regulator PhoP in Mycobacterium tuberculosis.
Sci Rep, 6, 2016
|
|
6X1T
 
 | | Structure of pHis Fab (SC50-3) in complex with pHis mimetic peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Wilson, I.A, Hunter, T. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ETF
 
 | |
3IDG
 
 | |
6UB7
 
 | | Crystal structure of a GH128 (subgroup V) exo-beta-1,3-glucanase from Cryptococcus neoformans (CnGH128_V) | | Descriptor: | Glyco_hydro_cc domain-containing protein, POTASSIUM ION | | Authors: | Santos, C.R, Costa, P.A.C.R, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
9ATM
 
 | | SARS-CoV-2 EG.5 RBD bound to the VIR-7229 and the S2H97 Fab fragments | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rietz, T, Park, Y.J, Errico, J, Czudnochowski, N, Nix, J.C, Corti, D, Snell, G, Marco, A.D, Pinto, D, Cameroni, E, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-02-27 | | Release date: | 2024-10-16 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 187, 2024
|
|
6BBV
 
 | | Crystal Structure of JAK2 in complex with compound 25 | | Descriptor: | N-{cis-3-[methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]cyclobutyl}propane-1-sulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Han, S. | | Deposit date: | 2017-10-19 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of N-{cis-3-[Methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]cyclobutyl}propane-1-sulfonamide (PF-04965842): A Selective JAK1 Clinical Candidate for the Treatment of Autoimmune Diseases.
J. Med. Chem., 61, 2018
|
|
