2F6C
 
 | | Reaction geometry and thermostability of pyranose 2-oxidase from the white-rot fungus Peniophora sp., Thermostability mutant E542K | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, Pyranose 2-oxidase, ... | | Authors: | Bannwarth, M, Heckmann-Pohl, D.M, Bastian, S, Giffhorn, F, Schulz, G.E. | | Deposit date: | 2005-11-29 | | Release date: | 2006-06-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Reaction Geometry and Thermostable Variant of Pyranose 2-Oxidase from the White-Rot Fungus Peniophora sp.
Biochemistry, 45, 2006
|
|
8S1K
 
 | | Crystal Structure of human FABP4 in complex with 2-[1-(methoxymethyl)cyclopentyl]-6-pentyl-4-phenyl-3-(1H-tetrazol-5-yl)-5,6,7,8-tetrahydroquinoline | | Descriptor: | (6~{S})-2-[1-(methoxymethyl)cyclopentyl]-6-pentyl-4-phenyl-3-(1~{H}-1,2,3,4-tetrazol-5-yl)-5,6,7,8-tetrahydroquinoline, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2024-02-15 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
2CCE
 
 | | Parallel Configuration of pLI E20S | | Descriptor: | General control protein GCN4 | | Authors: | Yadav, M.K, Leman, L.J, Price, D.J, Brooks 3rd, C.L, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2006-01-16 | | Release date: | 2006-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Coiled coils at the edge of configurational heterogeneity. Structural analyses of parallel and antiparallel homotetrameric coiled coils reveal configurational sensitivity to a single solvent-exposed amino acid substitution.
Biochemistry, 45, 2006
|
|
2CCN
 
 | | pLI E20C is antiparallel | | Descriptor: | General control protein GCN4 | | Authors: | Yadav, M.K, Leman, L.J, Price, D.J, Brooks 3rd, C.L, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2006-01-16 | | Release date: | 2006-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Coiled coils at the edge of configurational heterogeneity. Structural analyses of parallel and antiparallel homotetrameric coiled coils reveal configurational sensitivity to a single solvent-exposed amino acid substitution.
Biochemistry, 45, 2006
|
|
2CCF
 
 | | Antiparallel Configuration of pLI E20S | | Descriptor: | General control protein GCN4 | | Authors: | Yadav, M.K, Leman, L.J, Price, D.J, Brooks 3rd, C.L, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2006-01-16 | | Release date: | 2006-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Coiled coils at the edge of configurational heterogeneity. Structural analyses of parallel and antiparallel homotetrameric coiled coils reveal configurational sensitivity to a single solvent-exposed amino acid substitution.
Biochemistry, 45, 2006
|
|
4ZXG
 
 | | Ligandin binding site of PfGST | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Glutathione S-transferase, ... | | Authors: | Perbandt, M, Eberle, R, Betzel, C. | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution structures of Plasmodium falciparum GST complexes provide novel insights into the dimer-tetramer transition and a novel ligand-binding site.
J.Struct.Biol., 191, 2015
|
|
2FUB
 
 | | Crystal structure of urate oxidase at 140 MPa | | Descriptor: | 8-AZAXANTHINE, CYSTEINE, Uricase | | Authors: | Colloc'h, N, Girard, E, Fourme, R. | | Deposit date: | 2006-01-26 | | Release date: | 2006-02-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High pressure macromolecular crystallography: The 140-MPa crystal structure at 2.3 A resolution of urate oxidase, a 135-kDa tetrameric assembly
Biochim.Biophys.Acta, 1764, 2006
|
|
7FX6
 
 | | Crystal Structure of human FABP4 in complex with N,N-diethyl-4-pyridin-4-yl-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridin-2-amine | | Descriptor: | (3M)-N,N-diethyl-4-(pyridin-4-yl)-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridin-2-amine, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
7G1A
 
 | | Crystal Structure of human FABP4 in complex with 2-[(3S)-oxolan-3-yl]oxy-4-phenyl-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridine | | Descriptor: | (3M)-2-{[(3S)-oxolan-3-yl]oxy}-4-phenyl-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridine, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Buettelmann, B, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
7FX8
 
 | | Crystal Structure of human FABP4 in complex with 4-(2-methylpropyl)-2-phenyl-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridine | | Descriptor: | (3M)-4-(2-methylpropyl)-2-phenyl-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridine, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
7FXH
 
 | | Crystal Structure of human FABP4 in complex with 2-[(2R)-oxolan-2-yl]-4-phenyl-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridine | | Descriptor: | (3P)-2-[(2R)-oxolan-2-yl]-4-phenyl-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridine, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
7FXN
 
 | | Crystal Structure of human FABP4 in complex with 2-[(2S)-oxolan-2-yl]-4-phenyl-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridine | | Descriptor: | (3P)-2-[(2S)-oxolan-2-yl]-4-phenyl-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridine, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
3I5A
 
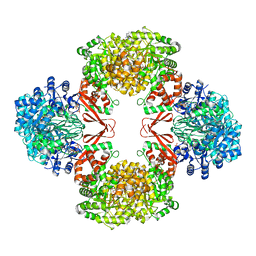
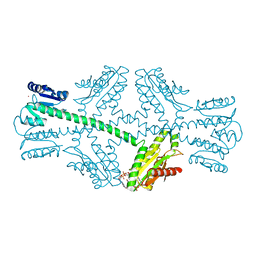 | | Crystal structure of full-length WpsR from Pseudomonas syringae | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Response regulator/GGDEF domain protein, STRONTIUM ION | | Authors: | Navarro, M.V.A.S, De, N, Sondermann, H. | | Deposit date: | 2009-07-03 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Determinants for the activation and autoinhibition of the diguanylate cyclase response regulator WspR.
J.Mol.Biol., 393, 2009
|
|
6RL5
 
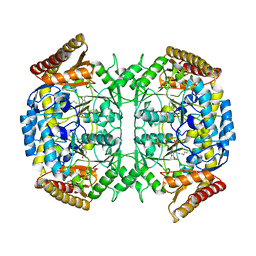 | |
7PT1
 
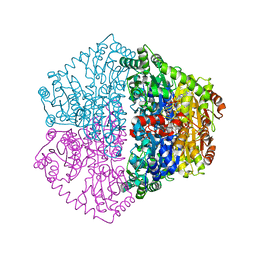 | |
7PT2
 
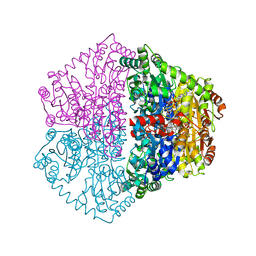 | |
7PT3
 
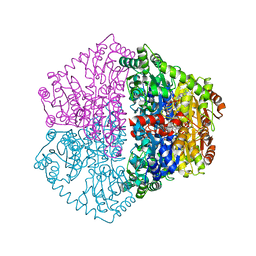 | |
7ZZ8
 
 | | Cryo-EM structure of Lactococcus lactis pyruvate carboxylase with acetyl-CoA and cyclic di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
1PLF
 
 | |
1YOD
 
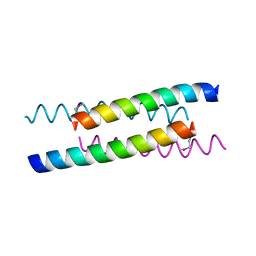 | |
7BZV
 
 | |
1VLO
 
 | |
1W5H
 
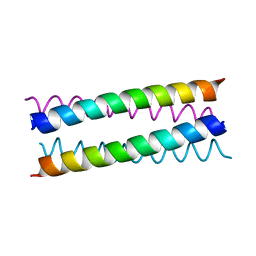 | | An anti-parallel four helix bundle. | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Leman, L.J, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2004-08-06 | | Release date: | 2004-09-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Coiled Coils at the Edge of Configurational Heterogeneity. Structural Analyses of Parallel and Antiparallel Homotetrameric Coiled Coils Reveal Configurational Sensitivity to a Single Solvent-Exposed Amino Acid Substitution.
Biochemistry, 45, 2006
|
|
5ZQU
 
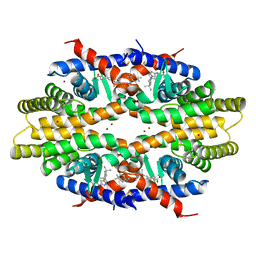 | | Crystal structure of tetrameric RXRalpha-LBD complexed with partial agonist CBt-PMN | | Descriptor: | 1-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)benzotriazole-5-carboxylic acid, BROMIDE ION, Retinoic acid receptor RXR-alpha | | Authors: | Miyashita, Y, Numoto, N, Arulmozhiraja, S, Nakano, S, Matsuo, N, Shimizu, K, Kakuta, H, Ito, S, Ikura, T, Ito, N, Tokiwa, H. | | Deposit date: | 2018-04-20 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.60038781 Å) | | Cite: | Dual conformation of the ligand induces the partial agonistic activity of retinoid X receptor alpha (RXR alpha ).
FEBS Lett., 593, 2019
|
|
6LPG
 
 | | human VASH1-SVBP complex | | Descriptor: | SULFATE ION, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Ikeda, A, Nishino, T. | | Deposit date: | 2020-01-10 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the tetrameric human vasohibin-1-SVBP complex reveals a variable arm region within the structural core.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
