3J77
 
 | | Structures of yeast 80S ribosome-tRNA complexes in the rotated and non-rotated conformations (Class II - rotated ribosome with 1 tRNA) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Svidritskiy, E, Brilot, A.F, Koh, C.S, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-05-29 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structures of Yeast 80S Ribosome-tRNA Complexes in the Rotated and Nonrotated Conformations.
Structure, 22, 2014
|
|
7NP7
 
 | | Structure of an intact ESX-5 inner membrane complex, Composite C1 model | | Descriptor: | ESX-5 secretion system ATPase EccB5, ESX-5 secretion system protein EccC5, ESX-5 secretion system protein EccD5, ... | | Authors: | Fahrenkamp, D, Bunduc, C.M, Wald, J, Ummels, R, Bitter, W, Houben, E.N.G, Marlovits, T.C. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structure and dynamics of a mycobacterial type VII secretion system.
Nature, 593, 2021
|
|
8ZFI
 
 | | Structure of E.coli ribosome in complex with an engineered arrest peptide and trigger factor | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Sriramoju, M.K, Ko, T.P, Draczkowski, P, Hsu, S.T.D. | | Deposit date: | 2024-05-07 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of E.coli ribosome in complex with an engineered arrest peptide and trigger factor
To Be Published
|
|
8ZEJ
 
 | | Structure of E.coli ribosome in complex with an engineered arrest peptide | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Sriramoju, M.K, Ko, T.P, Draczkowski, P, Hsu, S.T.D. | | Deposit date: | 2024-05-06 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure of E.coli ribosome in complex with an engineered arrest peptide
To Be Published
|
|
4LLG
 
 | | Crystal Structure Analysis of the E.coli holoenzyme/Gp2 complex | | Descriptor: | Bacterial RNA polymerase inhibitor, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2013-07-09 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.7936 Å) | | Cite: | Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma 70 domain 1.1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7N7G
 
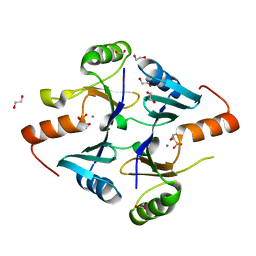 | | Crystal Structure of FosB from Enterococcus faecium with Fosfomycin | | Descriptor: | 1,2-ETHANEDIOL, FOSFOMYCIN, MANGANESE (II) ION, ... | | Authors: | Shay, M.R, Simmons, Z, Thompson, M.K. | | Deposit date: | 2021-06-10 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional characterization of fosfomycin resistance conferred by FosB from Enterococcus faecium.
Protein Sci., 31, 2022
|
|
4LJZ
 
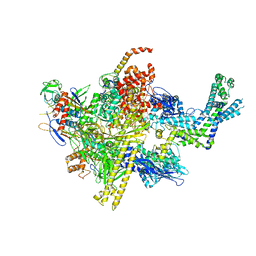 | | Crystal Structure Analysis of the E.coli holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2013-07-05 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5871 Å) | | Cite: | Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma 70 domain 1.1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
9BRB
 
 | |
9BRD
 
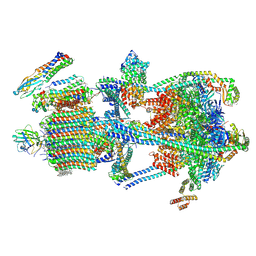 | | Synaptic Vesicle V-ATPase with synaptophysin and SidK, State 3 | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Coupland, C.E, Rubinstein, J.L. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles.
Science, 385, 2024
|
|
9BRC
 
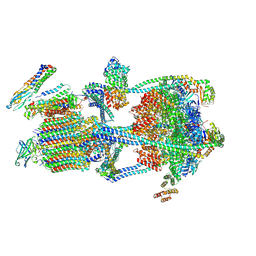 | |
4LK0
 
 | | Crystal Structure Analysis of the E.coli holoenzyme/T7 Gp2 complex | | Descriptor: | Bacterial RNA polymerase inhibitor, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2013-07-05 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.9103 Å) | | Cite: | Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma 70 domain 1.1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5J7L
 
 | | Structure of the 70S E coli ribosome with the U1052G mutation in the 16S rRNA bound to tetracycline | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-04-06 | | Release date: | 2016-07-27 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7NPS
 
 | | Structure of the periplasmic assembly from the ESX-5 inner membrane complex, C1 model | | Descriptor: | ESX-5 secretion system ATPase EccB5, Mycosin-5 | | Authors: | Fahrenkamp, D, Bunduc, C.M, Wald, J, Ummels, R, Bitter, W, Houben, E.N.G, Marlovits, T.C. | | Deposit date: | 2021-02-28 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structure and dynamics of a mycobacterial type VII secretion system.
Nature, 593, 2021
|
|
3JZ7
 
 | |
9BQ4
 
 | |
3JCU
 
 | | Cryo-EM structure of spinach PSII-LHCII supercomplex at 3.2 Angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Wei, X.P, Zhang, X.Z, Su, X.D, Cao, P, Liu, X.Y, Li, M, Chang, W.R, Liu, Z.F. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of spinach photosystem II-LHCII supercomplex at 3.2 A resolution
Nature, 534, 2016
|
|
3D23
 
 | | Main protease of HCoV-HKU1 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Zhao, Q, Chen, C, Li, S, Zou, Y. | | Deposit date: | 2008-05-07 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1.
J.Virol., 82, 2008
|
|
1KNK
 
 | | Crystal Structure of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate Synthase (ispF) from E. coli involved in Mevalonate-Independent Isoprenoid Biosynthesis | | Descriptor: | 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, MANGANESE (II) ION | | Authors: | Richard, S.B, Ferrer, J.L, Bowman, M.E, Lillo, A.M, Tetzlaff, C.N, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-12-18 | | Release date: | 2002-06-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase. An enzyme in the mevalonate-independent isoprenoid biosynthetic pathway.
J.Biol.Chem., 277, 2002
|
|
3D73
 
 | | Crystal structure of a pheromone binding protein mutant D35A, from Apis mellifera, at pH 7.0 | | Descriptor: | N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
3D78
 
 | | Dimeric crystal structure of a pheromone binding protein mutant D35N, from apis mellifera, at pH 7.0 | | Descriptor: | 1,2-ETHANEDIOL, N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
4LKU
 
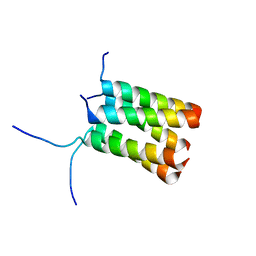 | |
3CTL
 
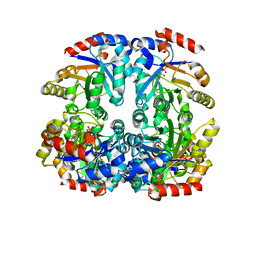 | | Crystal structure of D-Allulose 6-Phosphate 3-Epimerase from Escherichia coli K12 complexed with D-glucitol 6-phosphate and magnesium | | Descriptor: | D-SORBITOL-6-PHOSPHATE, D-allulose-6-phosphate 3-epimerase, MAGNESIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-04-14 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for substrate specificity in phosphate binding (beta/alpha)8-barrels: D-allulose 6-phosphate 3-epimerase from Escherichia coli K-12.
Biochemistry, 47, 2008
|
|
3JA1
 
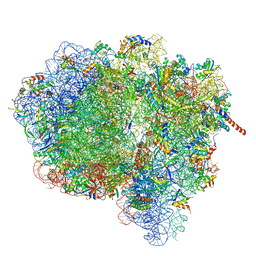 | | Activation of GTP Hydrolysis in mRNA-tRNA Translocation by Elongation Factor G | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Li, W, Liu, Z, Koripella, R.K, Langlois, R, Sanyal, S, Frank, J. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G.
Sci Adv, 1, 2015
|
|
9B8O
 
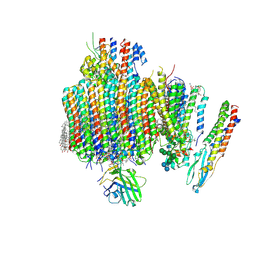 | | Synaptic Vesicle V-ATPase with synaptophysin and SidK, State 3, Vo | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Coupland, C.E, Rubinstein, J.L. | | Deposit date: | 2024-03-31 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles.
Science, 385, 2024
|
|
9L5R
 
 | | Cryo-EM structure of the thermophile spliceosome (state ILS) | | Descriptor: | Anaphase-promoting complex subunit 4-like WD40 domain-containing protein, CCDC12, Cell cycle control protein, ... | | Authors: | Li, Y, Fischer, P, Wang, M, Yuan, R, Meng, W, Luehrmann, R, Lau, B, Hurt, E, Cheng, J. | | Deposit date: | 2024-12-23 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into spliceosome fidelity: DHX35-GPATCH1- mediated rejection of aberrant splicing substrates.
Cell Res., 35, 2025
|
|
