5OES
 
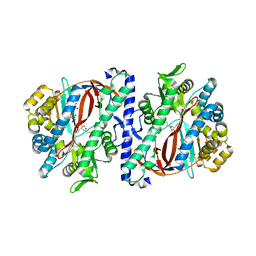 | | The structure of a glutathione synthetase (StGSS1) from Solanum tuberosum in ADP and y-EC bound closed conformation. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GAMMA-GLUTAMYLCYSTEINE, Glutathione synthetase, ... | | Authors: | Lilley, C.J, Maqbool, A, Wu, D, Yusup, H.B, Jones, L.M, Birch, P.R.J, Banfield, M.J, Urwin, P.E, Eves-van den Akker, S. | | Deposit date: | 2017-07-10 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Effector gene birth in plant parasitic nematodes: Neofunctionalization of a housekeeping glutathione synthetase gene.
PLoS Genet., 14, 2018
|
|
6WH1
 
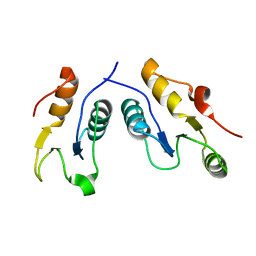 | | Structure of the complex of human DNA ligase III-alpha and XRCC1 BRCT domains | | Descriptor: | DNA ligase 3 alpha, X-ray repair cross complementing protein 1 variant | | Authors: | Pourfarjam, Y, Ellenberger, T, Tainer, J.A, Tomkinson, A.E, Kim, I.K. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase III alpha within a flexible DNA repair complex.
Nucleic Acids Res., 49, 2021
|
|
2E2W
 
 | |
7L8N
 
 | |
5ZJM
 
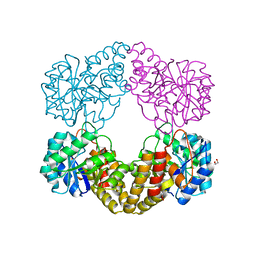 | | Crystal structure of N-acetylneuraminate lyase from Fusobacterium nucleatum | | Descriptor: | 1,2-ETHANEDIOL, N-acetylneuraminate lyase | | Authors: | Kumar, J.P, Rao, H, Nayak, V, Subramanian, R. | | Deposit date: | 2018-03-21 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.323 Å) | | Cite: | Crystal structures and kinetics of N-acetylneuraminate lyase from Fusobacterium nucleatum
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5U95
 
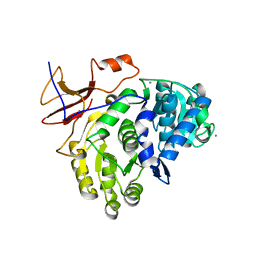 | |
4HR7
 
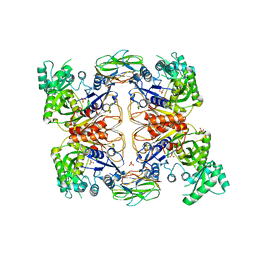 | | Crystal Structure of Biotin Carboxyl Carrier Protein-Biotin Carboxylase Complex from E.coli | | Descriptor: | 1,2-ETHANEDIOL, Biotin carboxyl carrier protein of acetyl-CoA carboxylase, Biotin carboxylase, ... | | Authors: | Broussard, T.C, Kobe, M.J, Pakhomova, S, Neau, D.B, Price, A.E, Champion, T.S, Waldrop, G.L. | | Deposit date: | 2012-10-26 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | The three-dimensional structure of the biotin carboxylase-biotin carboxyl carrier protein complex of E. coli acetyl-CoA carboxylase.
Structure, 21, 2013
|
|
4YS8
 
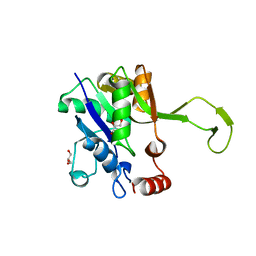 | |
4ZLK
 
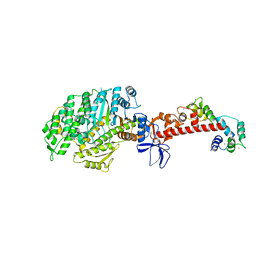 | | Crystal structure of mouse myosin-5a in complex with calcium-bound calmodulin | | Descriptor: | CALCIUM ION, Calmodulin, Unconventional myosin-Va | | Authors: | Shen, M, Zhang, N, Zheng, S, Zhang, W.-B, Zhang, H.-M, Lu, Z, Su, Q.P, Sun, Y, Ye, K, Li, X.-D. | | Deposit date: | 2015-05-01 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural basis for calcium regulation of myosin 5 motor function
To Be Published
|
|
4JLZ
 
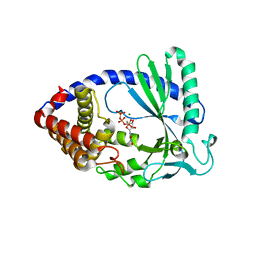 | |
8BIT
 
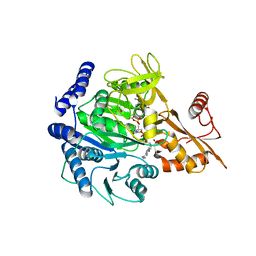 | | Crystal structure of acyl-CoA synthetase from Metallosphaera sedula in complex with Coenzyme A and acetyl-AMP | | Descriptor: | 4-hydroxybutyrate--CoA ligase 1, COENZYME A, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] ethanoate | | Authors: | Capra, N, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2022-11-02 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Adapting an acyl CoA ligase from Metallosphaera sedula for lactam formation by structure-guided protein engineering
Front Catal, 4, 2024
|
|
8BIQ
 
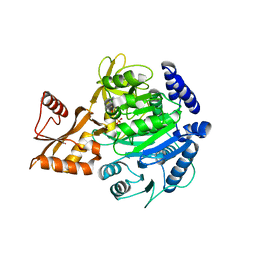 | | Crystal structure of acyl-COA synthetase from Metallosphaera sedula in complex with acetyl-AMP | | Descriptor: | 4-hydroxybutyrate--CoA ligase 1, ADENOSINE MONOPHOSPHATE, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] ethanoate | | Authors: | Capra, N, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2022-11-02 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Adapting an acyl CoA ligase from Metallosphaera sedula for lactam formation by structure-guided protein engineering
Front Catal, 4, 2024
|
|
2YL2
 
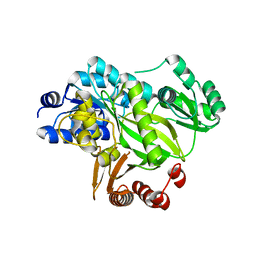 | | Crystal structure of human acetyl-CoA carboxylase 1, biotin carboxylase (BC) domain | | Descriptor: | ACETYL-COA CARBOXYLASE 1 | | Authors: | Muniz, J.R.C, Froese, D.S, Krysztofinska, E, Vollmar, M, Beltrami, A, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Yue, W.W, Oppermann, U. | | Deposit date: | 2011-05-31 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Human Acetyl-Coa Carboxylase 1, Biotin Carboxylase (Bc) Domain
To be Published
|
|
2ZW2
 
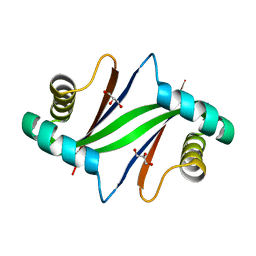 | | Crystal Structure of Formylglycinamide Ribonucleotide Amidotransferase III from SULFOLOBUS TOKODAII (STPURS) | | Descriptor: | GLYCEROL, Putative uncharacterized protein STS178 | | Authors: | Suzuki, S, Tamura, S, Okada, K, Baba, S, Kumasaka, T, Nakagawa, N, Masui, R, Kuramitsu, S, Sampei, G, Kawai, G. | | Deposit date: | 2008-11-27 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Formylglycinamide Ribonucleotide Amidotransferase III from SULFOLOBUS TOKODAII (STPURS)
To be Published
|
|
5CSK
 
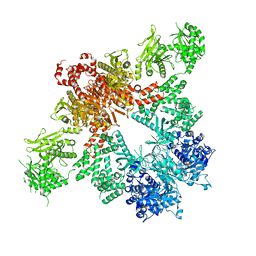 | |
6ANU
 
 | | Cryo-EM structure of F-actin complexed with the beta-III-spectrin actin-binding domain | | Descriptor: | Actin, cytoplasmic 1, Spectrin beta chain, ... | | Authors: | Wang, F, Orlova, A, Avery, A.W, Hays, T.S, Egelman, E.H. | | Deposit date: | 2017-08-14 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis for high-affinity actin binding revealed by a beta-III-spectrin SCA5 missense mutation.
Nat Commun, 8, 2017
|
|
6V99
 
 | | Agrobacterium tumefaciens ADP-Glucose pyrophosphorylase- S72D in the presence of sulfate | | Descriptor: | Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Zheng, Y, Alghamdi, M.A, Ballicora, M.A, Liu, D. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Site-directed mutagenesis of Serine-72 reveals the location of the fructose 6-phosphate regulatory site of the Agrobacterium tumefaciens ADP-glucose pyrophosphorylase.
Protein Sci., 31, 2022
|
|
8F41
 
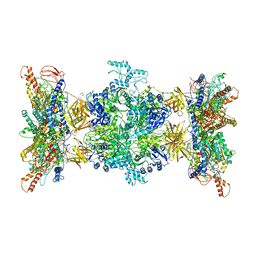 | | 3-methylcrotonyl-CoA carboxylase in filament, alpha-subunit centered | | Descriptor: | 3-methylcrotonyl-CoA carboxylase, alpha-subunit, beta-subunit, ... | | Authors: | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-11-10 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
8F3D
 
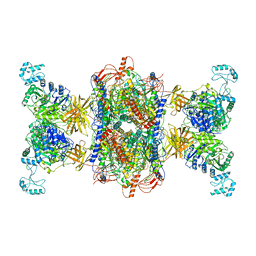 | | 3-methylcrotonyl-CoA carboxylase in filament, beta-subunit centered | | Descriptor: | 3-methylcrotonyl-CoA carboxylase alpha-subunit, 3-methylcrotonyl-CoA carboxylase beta-subunit, 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL | | Authors: | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-11-09 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
6V9A
 
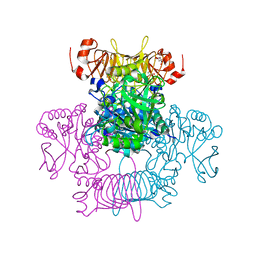 | | Agrobacterium tumefaciens ADP-Glucose pyrophosphorylase-S72D | | Descriptor: | CITRIC ACID, GLYCEROL, Glucose-1-phosphate adenylyltransferase | | Authors: | Zheng, Y, Alghamdi, M.A, Ballicora, M.A, Liu, D. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Site-directed mutagenesis of Serine-72 reveals the location of the fructose 6-phosphate regulatory site of the Agrobacterium tumefaciens ADP-glucose pyrophosphorylase.
Protein Sci., 31, 2022
|
|
6V96
 
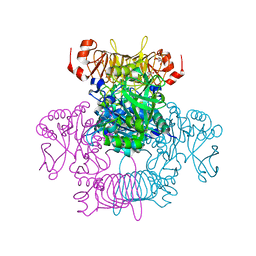 | | Agrobacterium tumefaciens ADP-Glucose pyrophosphorylase-S72E | | Descriptor: | CITRIC ACID, GLYCEROL, Glucose-1-phosphate adenylyltransferase | | Authors: | Zheng, Y, Hussien, R, Alghamdi, M.A, Ballicora, M.A, Liu, D. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Site-directed mutagenesis of Serine-72 reveals the location of the fructose 6-phosphate regulatory site of the Agrobacterium tumefaciens ADP-glucose pyrophosphorylase.
Protein Sci., 31, 2022
|
|
4Q16
 
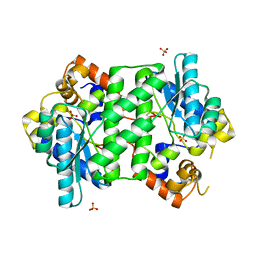 | |
3N05
 
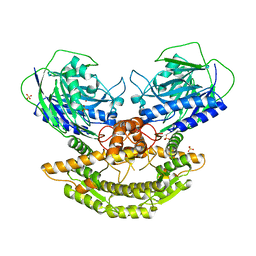 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM STREPTOMYCES AVERMITILIS | | Descriptor: | NH(3)-dependent NAD(+) synthetase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Do, J, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-13 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Nh3-Dependent Nad+ Synthetase from Streptomyces Avermitilis
To be Published
|
|
3DPI
 
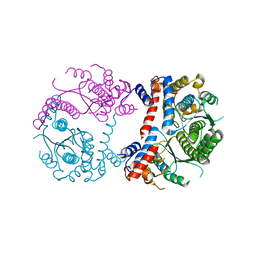 | |
3N6R
 
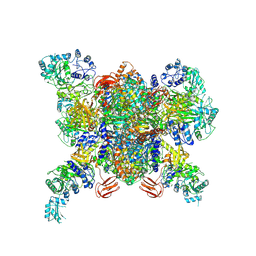 | | CRYSTAL STRUCTURE OF the holoenzyme of PROPIONYL-COA CARBOXYLASE (PCC) | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Propionyl-CoA carboxylase, alpha subunit, ... | | Authors: | Huang, C.S, Sadre-Bazzaz, K, Tong, L. | | Deposit date: | 2010-05-26 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the alpha(6)beta(6) holoenzyme of propionyl-coenzyme A carboxylase.
Nature, 466, 2010
|
|
