4PBR
 
 | | Crystal structure of the M. jannaschii G2 tRNA synthetase variant bound to 4-(2-bromoisobutyramido)-phenylalanine (BibaF) | | Descriptor: | 4-[(2-bromo-2-methylpropanoyl)amino]-L-phenylalanine, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Cooley, R.B, Karplus, P.A, Mehl, R.A. | | Deposit date: | 2014-04-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gleaning Unexpected Fruits from Hard-Won Synthetases: Probing Principles of Permissivity in Non-canonical Amino Acid-tRNA Synthetases.
Chembiochem, 15, 2014
|
|
7BZJ
 
 | | The Discovery of Benzhydrol-Oxaborole Hybrid Derivatives as Leucyl-tRNA Synthetase Inhibitors | | Descriptor: | Leucine--tRNA ligase, [(1~{R},5~{R},6~{S},8~{R})-8-(6-aminopurin-9-yl)-4'-[(~{R})-oxidanyl-[4-(2-oxidanylidenepropylsulfanyl)phenyl]methyl]spiro[2,4,7-trioxa-3-boranuidabicyclo[3.3.0]octane-3,7'-7-boranuidabicyclo[4.3.0]nona-1(6),2,4-triene]-6-yl]methoxy-tris(oxidanyl)phosphanium | | Authors: | Liu, R.J, Li, H, Wang, E.D, Zhou, H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of benzhydrol-oxaborole derivatives as Streptococcus pneumoniae leucyl-tRNA synthetase inhibitors.
Bioorg.Med.Chem., 29, 2021
|
|
1JD9
 
 | |
2JEM
 
 | | Native family 12 xyloglucanase from Bacillus licheniformis | | Descriptor: | ENDO-BETA-1,4-GLUCANASE | | Authors: | Gloster, T.M, Ibatullin, F.M, Macauley, K, Eklof, J.M, Roberts, S, Turkenburg, J.P, Bjornvad, M.E, Jorgensen, P.L, Danielsen, S, Johansen, K.S, Borchert, T.V, Wilson, K.S, Brumer, H, Davies, G.J. | | Deposit date: | 2007-01-18 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Characterization and Three-Dimensional Structures of Two Distinct Bacterial Xyloglucanases from Families Gh5 and Gh12.
J.Biol.Chem., 282, 2007
|
|
8V2W
 
 | | Crystal Structure of the ancestral triosephosphate isomerase reconstruction of the last opisthokont common ancestor obtained by Bayesian inference | | Descriptor: | GLYCEROL, Triosephosphate isomerase | | Authors: | Perez-Nino, J.A, Rodriguez-Romero, A, Guerra, Y, Fernandez-Velasco, D.A. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Protein Sci., 33, 2024
|
|
8W08
 
 | | Crystal Structure of the worst case reconstruction of the ancestral triosephosphate isomerase of the last opisthokont common ancestor obtained by maximum likelihood | | Descriptor: | FLUORIDE ION, Triosephosphate isomerase | | Authors: | Perez-Nino, J.A, Rodriguez-Romero, A, Guerra-Borrego, Y, Fernandez-Velasco, D.A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Protein Sci., 33, 2024
|
|
8V0A
 
 | | Crystal Structure of the worst case of the reconstruction of the ancestral triosephosphate isomerase of the last opisthokont common ancestor obtained by maximum likelihood with PGH | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, Triosephosphate isomerase | | Authors: | Perez-Nino, J.A, Rodriguez-Romero, A, Guerra-Borrego, Y, Fernandez-Velasco, D.A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Protein Sci., 33, 2024
|
|
8W05
 
 | |
4L5V
 
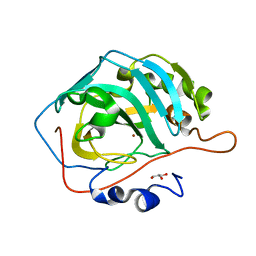 | | The structural implications of the secondary CO2 binding pocket in human carbonic anhydrase II | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, ZINC ION | | Authors: | Boone, C.D, Gill, S, McKenna, R. | | Deposit date: | 2013-06-11 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural, catalytic and stabilizing consequences of aromatic cluster variants in human carbonic anhydrase II.
Arch.Biochem.Biophys., 539, 2013
|
|
6QTM
 
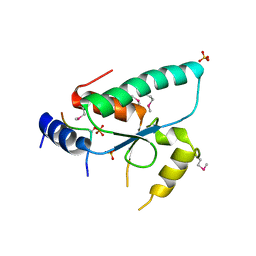 | | Crystal structure of the Sir4 H-BRCT domain in complex with Ty5 pS1095 peptide | | Descriptor: | Regulatory protein SIR4, Ribonuclease H, SULFATE ION | | Authors: | Gut, H, Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M. | | Deposit date: | 2019-02-25 | | Release date: | 2019-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
5EPL
 
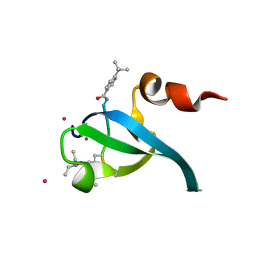 | | Crystal Structure of chromodomain of CBX4 in complex with inhibitor UNC3866 | | Descriptor: | E3 SUMO-protein ligase CBX4, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
6DQN
 
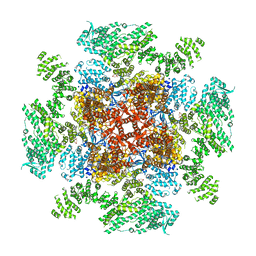 | | Class 1 IP3-bound human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DQZ
 
 | | Class 4 IP3-bound human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.01 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5EAR
 
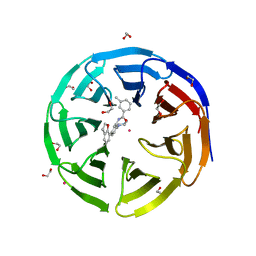 | | Crystal structure of human WDR5 in complex with compound 9d | | Descriptor: | 1,2-ETHANEDIOL, N-[5-(2,3-dihydro-1-benzofuran-7-yl)-2-(4-methylpiperazin-1-yl)phenyl]-3-methylbenzamide, SULFATE ION, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1).
J. Med. Chem., 59, 2016
|
|
3O1D
 
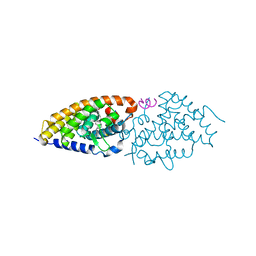 | | Structure-function study of Gemini derivatives with two different side chains at C-20, Gemini-0072 and Gemini-0097. | | Descriptor: | (1R,3R,7E,17beta)-17-[(1S)-6,6,6-trifluoro-5-hydroxy-1-(4-hydroxy-4-methylpentyl)-5-(trifluoromethyl)hex-3-yn-1-yl]-9,10-secoestra-5,7-diene-1,3-diol, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | Authors: | Huet, T, Moras, D, Rochel, N. | | Deposit date: | 2010-07-21 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4007 Å) | | Cite: | Structure-function study of gemini derivatives with two different side chains at C-20, Gemini-0072 and Gemini-0097.
Medchemcomm, 2, 2011
|
|
5DN4
 
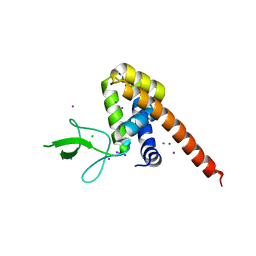 | |
1PNC
 
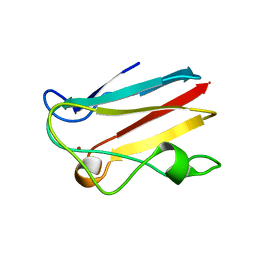 | |
3ILY
 
 | |
5E17
 
 | |
8D39
 
 | |
4HVC
 
 | | Crystal structure of human prolyl-tRNA synthetase in complex with halofuginone and ATP analogue | | Descriptor: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, Bifunctional glutamate/proline--tRNA ligase, MAGNESIUM ION, ... | | Authors: | Zhou, H, Sun, L, Yang, X.L, Schimmel, P. | | Deposit date: | 2012-11-06 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ATP-directed capture of bioactive herbal-based medicine on human tRNA synthetase.
Nature, 494, 2012
|
|
4QH1
 
 | | The GLIC pentameric Ligand-Gated Ion Channel (wild-type) in complex with bromoacetate | | Descriptor: | BROMIDE ION, Proton-gated ion channel, SODIUM ION, ... | | Authors: | Fourati, Z, Delarue, M, Sauguet, L. | | Deposit date: | 2014-05-26 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural characterization of potential modulation sites in the extracellular domain of the prokaryotic pentameric proton-gated ion channel GLIC
Acta Crystallogr.,Sect.D, 2015
|
|
6ROT
 
 | | Thrombin in complex with MI2105 | | Descriptor: | (2~{S})-~{N}-[[5-chloranyl-2-(hydroxymethyl)phenyl]methyl]-1-[2-[(phenylmethyl)sulfonylamino]ethanoyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G. | | Deposit date: | 2019-05-13 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.339 Å) | | Cite: | Strategies for Late-Stage Optimization: Profiling Thermodynamics by Preorganization and Salt Bridge Shielding.
J.Med.Chem., 62, 2019
|
|
5AVH
 
 | | The 0.90 angstrom structure (I222) of glucose isomerase crystallized in high-strength agarose hydrogel | | Descriptor: | Xylose isomerase | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, N, Sazaki, G, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
4HWE
 
 | |
