6BJY
 
 | | VSV Nucleocapsid with Polyamide Bound | | Descriptor: | 4-{[4-(acetylamino)-1-methyl-1H-pyrrole-2-carbonyl]amino}-1-methyl-N-{4-[(1-methyl-1H-pyrrol-3-yl)amino]-4-oxobutyl}-1H-imidazole-2-carboxamide, Nucleoprotein, RNA (45-MER), ... | | Authors: | Gumpper, R.H, Luo, M. | | Deposit date: | 2017-11-07 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | A Polyamide Inhibits Replication of Vesicular Stomatitis Virus by Targeting RNA in the Nucleocapsid.
J. Virol., 92, 2018
|
|
6BS5
 
 | | Crystal structure of AMP-PNP-bound bacterial Get3-like A and B in Mycobacterium tuberculosis | | Descriptor: | Anion transporter, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, H, Hu, K, Kovach, A. | | Deposit date: | 2017-12-01 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Characterization of Guided Entry of Tail-Anchored Proteins 3 Homologues in Mycobacterium tuberculosis.
J.Bacteriol., 201, 2019
|
|
7K14
 
 | | Ternary soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN and methanesulfonate | | Descriptor: | Alkanesulfonate monooxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liew, J.J.M, Dowling, D.P, El Saudi, I.M. | | Deposit date: | 2020-09-07 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7K64
 
 | | Binary titrated soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN | | Descriptor: | Alkanesulfonate monooxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liew, J.J.M, Dowling, D.P. | | Deposit date: | 2020-09-18 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
6DAW
 
 | | X-ray crystal structure of NapI L-arginine desaturase bound to Fe(II), L-arginine, and acetate | | Descriptor: | ACETATE ION, ARGININE, FE (II) ION, ... | | Authors: | Mitchell, A.J, Dunham, N.P, Boal, A.K. | | Deposit date: | 2018-05-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two Distinct Mechanisms for C-C Desaturation by Iron(II)- and 2-(Oxo)glutarate-Dependent Oxygenases: Importance of alpha-Heteroatom Assistance.
J. Am. Chem. Soc., 140, 2018
|
|
6DB2
 
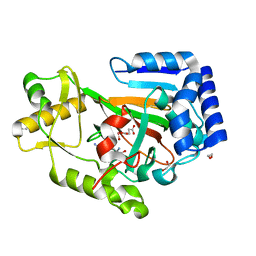 | | X-ray crystal structure of VioC bound to vanadyl ion, L-homoarginine, and succinate | | Descriptor: | 1,2-ETHANEDIOL, Alpha-ketoglutarate-dependent L-arginine hydroxylase, L-HOMOARGININE, ... | | Authors: | Dunham, N.P, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2018-05-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two Distinct Mechanisms for C-C Desaturation by Iron(II)- and 2-(Oxo)glutarate-Dependent Oxygenases: Importance of alpha-Heteroatom Assistance.
J. Am. Chem. Soc., 140, 2018
|
|
6CUM
 
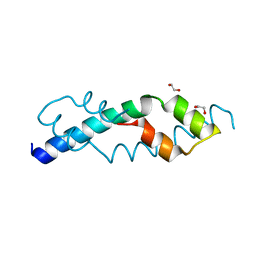 | |
6DHL
 
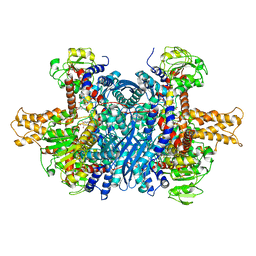 | | Bovine glutamate dehydrogenase complexed with epicatechin-3-gallate (ECG) | | Descriptor: | (2R,3S)-2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Smith, T.J. | | Deposit date: | 2018-05-20 | | Release date: | 2018-07-25 | | Method: | X-RAY DIFFRACTION (3.624 Å) | | Cite: | Green tea polyphenols control dysregulated glutamate dehydrogenase in transgenic mice by hijacking the ADP activation site.
J. Biol. Chem., 286, 2011
|
|
6DGO
 
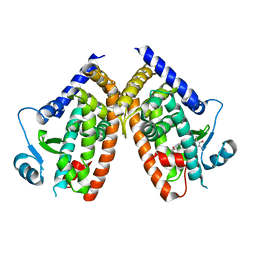 | | Crystal Structure of Human PPARgamma Ligand Binding Domain in Complex with Troglitazone | | Descriptor: | (5S)-5-[(4-{[(2R)-6-hydroxy-2,5,7,8-tetramethyl-3,4-dihydro-2H-1-benzopyran-2-yl]methoxy}phenyl)methyl]-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | Authors: | Shang, J, Kojetin, D.J. | | Deposit date: | 2018-05-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Quantitative structural assessment of graded receptor agonism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7L9X
 
 | | Structure of VPS4B in complex with an allele-specific covalent inhibitor | | Descriptor: | N-{3-[(8-phenyl[1,2,4]triazolo[1,5-a]pyridin-2-yl)amino]phenyl}propanamide, SULFATE ION, Vacuolar protein sorting-associated protein 4B | | Authors: | Grasso, M, Cupido, T, Kapoor, T.M. | | Deposit date: | 2021-01-05 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | A chemical genetics approach to examine the functions of AAA proteins.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6DAZ
 
 | | X-ray crystal structure of VioC bound to Fe(II), 3S-hydroxy-L-homoarginine, and succinate | | Descriptor: | (3S)-N~6~-carbamimidoyl-3-hydroxy-L-lysine, Alpha-ketoglutarate-dependent L-arginine hydroxylase, FE (II) ION, ... | | Authors: | Dunham, N.P, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2018-05-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Two Distinct Mechanisms for C-C Desaturation by Iron(II)- and 2-(Oxo)glutarate-Dependent Oxygenases: Importance of alpha-Heteroatom Assistance.
J. Am. Chem. Soc., 140, 2018
|
|
7MHU
 
 | | Sialidase24 apo | | Descriptor: | Exo-alpha-sialidase | | Authors: | Rees, S.D, Chang, G.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational models in the service of X-ray and cryo-electron microscopy structure determination.
Proteins, 89, 2021
|
|
6DHI
 
 | |
6DGL
 
 | |
6DAX
 
 | | X-ray crystal structure of VioC bound to Fe(II), L-homoarginine, and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent L-arginine hydroxylase, FE (II) ION, ... | | Authors: | Dunham, N.P, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2018-05-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two Distinct Mechanisms for C-C Desaturation by Iron(II)- and 2-(Oxo)glutarate-Dependent Oxygenases: Importance of alpha-Heteroatom Assistance.
J. Am. Chem. Soc., 140, 2018
|
|
6DGR
 
 | |
3IMX
 
 | | Crystal Structure of human glucokinase in complex with a synthetic activator | | Descriptor: | (2R)-3-cyclopentyl-N-(5-methoxy[1,3]thiazolo[5,4-b]pyridin-2-yl)-2-{4-[(4-methylpiperazin-1-yl)sulfonyl]phenyl}propanamide, Glucokinase, SODIUM ION, ... | | Authors: | Stams, T, Vash, B. | | Deposit date: | 2009-08-11 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigation of functionally liver selective glucokinase activators for the treatment of type 2 diabetes.
J.Med.Chem., 52, 2009
|
|
3IT3
 
 | | Crystal Structure Francisella tularensis histidine acid phosphatase D261A mutant complexed with substrate 3'-AMP | | Descriptor: | Acid phosphatase, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate | | Authors: | Singh, H, Felts, R.L, Reilly, T.J, Tanner, J.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of the histidine acid phosphatase from Francisella tularensis provide insight into substrate recognition.
J.Mol.Biol., 394, 2009
|
|
3IT0
 
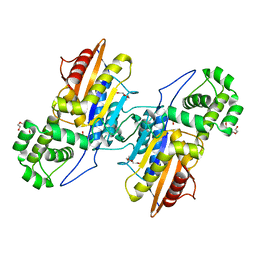 | | Crystal Structure Francisella tularensis histidine acid phosphatase complexed with phosphate | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Acid phosphatase, PENTAETHYLENE GLYCOL, ... | | Authors: | Singh, H, Felts, R.L, Reilly, T.J, Tanner, J.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Crystal Structures of the histidine acid phosphatase from Francisella tularensis provide insight into substrate recognition.
J.Mol.Biol., 394, 2009
|
|
3IT2
 
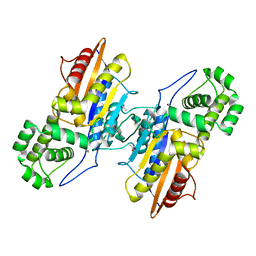 | | Crystal structure of ligand-free Francisella tularensis histidine acid phosphatase | | Descriptor: | ACETATE ION, Acid phosphatase | | Authors: | Singh, H, Felts, R.L, Reilly, T.J, Tanner, J.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | Crystal Structures of the histidine acid phosphatase from Francisella tularensis provide insight into substrate recognition.
J.Mol.Biol., 394, 2009
|
|
3KD5
 
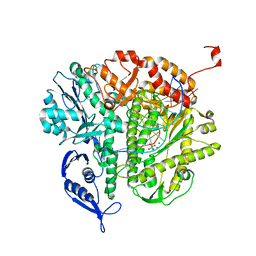 | |
3JCI
 
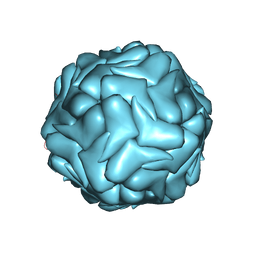 | | 2.9 Angstrom Resolution Cryo-EM 3-D Reconstruction of Close-packed PCV2 Virus-like Particles | | Descriptor: | Capsid protein | | Authors: | Liu, Z, Guo, F, Wang, F, Li, T.C, Jiang, W. | | Deposit date: | 2015-12-13 | | Release date: | 2016-02-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | 2.9 angstrom Resolution Cryo-EM 3D Reconstruction of Close-Packed Virus Particles.
Structure, 24, 2016
|
|
6T4C
 
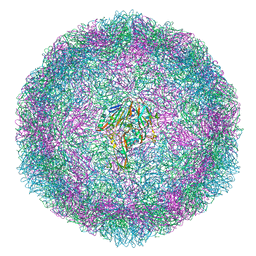 | | Bovine enterovirus F3 in complex with glutathione | | Descriptor: | GLUTATHIONE, GLYCEROL, POTASSIUM ION, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Walter, T.S, Fry, E.E, Stuart, D.I. | | Deposit date: | 2019-10-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glutathione facilitates enterovirus assembly by binding at a druggable pocket.
Commun Biol, 3, 2020
|
|
6TPI
 
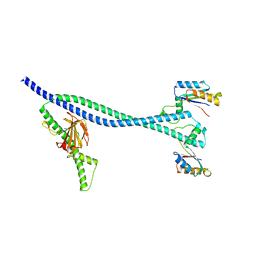 | | EnvC bound to the FtsX periplasmic domain | | Descriptor: | Cell division protein FtsX, Murein hydrolase activator EnvC | | Authors: | Crow, A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into bacterial cell division from a structure of EnvC bound to the FtsX periplasmic domain.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3KDK
 
 | |
