8V8D
 
 | | Alpha7-nicotinic acetylcholine receptor time resolved bound to epibatidine and PNU-120596 asymmetric state 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Burke, S.M, Noviello, C.M, Hibbs, R.E. | | Deposit date: | 2023-12-05 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
8V8C
 
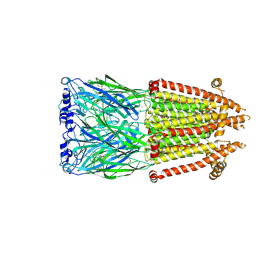 | | Alpha7-nicotinic acetylcholine receptor time resolved bound to epibatidine and PNU-120596 asymmetric state 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Burke, S.M, Noviello, C.M, Hibbs, R.E. | | Deposit date: | 2023-12-05 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
8V8B
 
 | |
8V8A
 
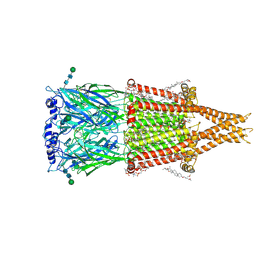 | | Alpha7-nicotinic acetylcholine receptor time resolved bound to epibatidine and PNU-120596 desensitized intermediate state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Burke, S.M, Noviello, C.M, Hibbs, R.E. | | Deposit date: | 2023-12-05 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
8V89
 
 | | Alpha7-nicotinic acetylcholine receptor time resolved resting state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Burke, S.M, Noviello, C.M, Hibbs, R.E. | | Deposit date: | 2023-12-05 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
8V88
 
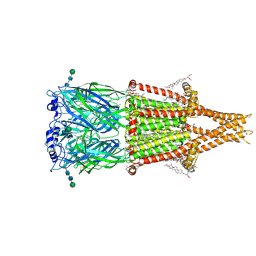 | | Alpha7-nicotinic acetylcholine receptor bound to epibatidine and GAT107 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (3aR,4S,9bS)-4-(4-bromophenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, ... | | Authors: | Burke, S.M, Noviello, C.M, Hibbs, R.E. | | Deposit date: | 2023-12-05 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
8V87
 
 | | 60S ribosome biogenesis intermediate (Dbp10 post-catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8V86
 
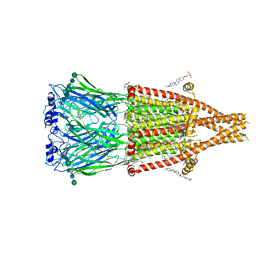 | | Alpha7-nicotinic acetylcholine receptor bound to GAT107 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (3aR,4S,9bS)-4-(4-bromophenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, ... | | Authors: | Burke, S.M, Noviello, C.M, Hibbs, R.E. | | Deposit date: | 2023-12-04 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
8V85
 
 | |
8V84
 
 | | 60S ribosome biogenesis intermediate (Dbp10 catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8V83
 
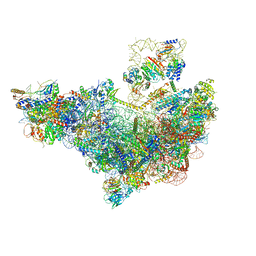 | | 60S ribosome biogenesis intermediate (Dbp10 pre-catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8V82
 
 | | Alpha7-nicotinic acetylcholine receptor bound to epibatidine and PNU-120596 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Burke, S.M, Noviello, C.M, Hibbs, R.E. | | Deposit date: | 2023-12-04 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
8V81
 
 | | Phosphorylated, ATP-bound, inhibitor 172-bound E1371Q human cystic fibrosis transmembrane conductance regulator | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 4-[(Z)-{(3M)-4-oxo-2-sulfanylidene-3-[3-(trifluoromethyl)phenyl]-1,3-thiazolidin-5-ylidene}methyl]benzoic acid, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gao, X, Hwang, T. | | Deposit date: | 2023-12-04 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Allosteric inhibition of CFTR gating by CFTRinh-172 binding in the pore
Nat Commun, 2024
|
|
8V80
 
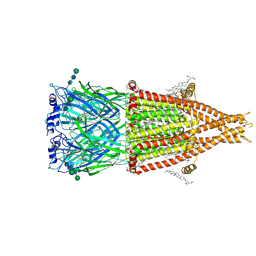 | | Alpha7-nicotinic acetylcholine receptor bound to epibatidine and (-)-TQS | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (3aR,4S,9bS)-4-(naphthalen-1-yl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, ... | | Authors: | Burke, S.M, Noviello, C.M, Hibbs, R.E. | | Deposit date: | 2023-12-04 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
8V7Z
 
 | |
8V7W
 
 | |
8V7U
 
 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z729726784 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclopentyl-N-(3-methyl-1,2,4-oxadiazol-5-yl)acetamide, DIMETHYL SULFOXIDE, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z729726784
To Be Published
|
|
8V7T
 
 | |
8V7R
 
 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z56772132 | | Descriptor: | (5R)-5-[2-(4-methoxyphenyl)ethyl]-5-methylimidazolidine-2,4-dione, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z56772132
To Be Published
|
|
8V7O
 
 | |
8V7L
 
 | | Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Histone H2A type 1, Histone H2B, ... | | Authors: | Chio, U.S, Palovcak, E, Armache, J.P, Narlikar, G.J, Cheng, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking.
Nat Commun, 15, 2024
|
|
8V7K
 
 | |
8V7J
 
 | |
8V7I
 
 | |
8V7H
 
 | |
