6X3R
 
 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, methyl (3~{S})-1-[(2~{S})-2-[[(2~{S})-2-acetamido-3-methyl-butanoyl]amino]-3-(3-hydroxyphenyl)propanoyl]-1,2-diazinane-3-carboxylate | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-21 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
7OAL
 
 | | Crystal structure of pseudokinase CASK in complex with compound 25 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[2,5-bis(bromanyl)-4-methyl-phenyl]methylamino]-4-(cyclohexylamino)-N-[3-(2-oxidanylidene-1,3-oxazolidin-3-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Russ, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
8PVU
 
 | |
7S6U
 
 | |
8C19
 
 | | SARS-CoV-2 NSP3 macrodomain in complex with 1-methyl-4-[5-(morpholin-4-ylcarbonyl)-2-furyl]-1H-pyrrolo[2,3-b]pyridine | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 3, [5-(1-methylpyrrolo[2,3-b]pyridin-4-yl)furan-2-yl]-morpholin-4-yl-methanone | | Authors: | Schuller, M, Ahel, I. | | Deposit date: | 2022-12-20 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Development Strategies for SARS-CoV-2 NSP3 Macrodomain Inhibitors.
Pathogens, 12, 2023
|
|
7MYI
 
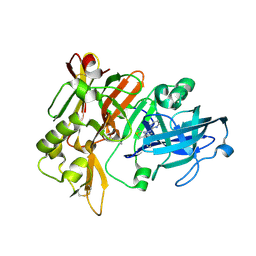 | | BACE-1 in complex with compound #6 | | Descriptor: | (4aR,7aR)-6-(pyrimidin-2-yl)-7a-(thiophen-2-yl)-4,4a,5,6,7,7a-hexahydropyrrolo[3,4-d][1,3]thiazin-2-amine, Beta-secretase 1, GLYCEROL | | Authors: | Hendle, J, Timm, D.E, Stout, S.L. | | Deposit date: | 2021-05-21 | | Release date: | 2021-07-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery and Early Clinical Development of LY3202626, a Low-Dose, CNS-Penetrant BACE Inhibitor.
J.Med.Chem., 64, 2021
|
|
6UR3
 
 | |
8BJ1
 
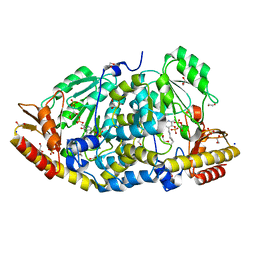 | | Crystal structure of Medicago truncatula histidinol-phosphate aminotransferase (HISN6) in the open state | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, SULFATE ION, ... | | Authors: | Rutkiewicz, M, Ruszkowski, M. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Insights into the substrate specificity, structure, and dynamics of plant histidinol-phosphate aminotransferase (HISN6).
Plant Physiol Biochem., 196, 2023
|
|
8Y5U
 
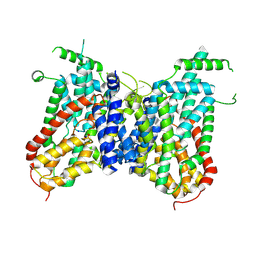 | | human NaS1 intermediate state 1 | | Descriptor: | SODIUM ION, SULFATE ION, Solute carrier family 13 member 1 | | Authors: | Zhang, S.S. | | Deposit date: | 2024-02-01 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | human NaS1 intermediate state 1
Sci Adv, 10, 2024
|
|
8F4A
 
 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with chlorhexidine | | Descriptor: | 1-[6-[azanylidene-[[azanylidene-[[(4-chlorophenyl)amino]methyl]-$l^{4}-azanyl]methyl]-$l^{4}-azanyl]hexyl]-3-[~{N}-(4-chlorophenyl)carbamimidoyl]guanidine, GLYCEROL, N-acetyltransferase Eis | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-10 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
8OIJ
 
 | | Drosophila Smaug-Smoothened complex | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Protein Smaug, ... | | Authors: | Ubartaite, G, Kubikova, J, Jeske, M. | | Deposit date: | 2023-03-23 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for binding of Drosophila Smaug to the GPCR Smoothened and to the germline inducer Oskar.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EQU
 
 | | Structure of SARS-CoV-2 Orf3a in late endosome/lysosome-like environment, Saposin A nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ORF3a protein, Saposin A, ... | | Authors: | Miller, A.N, Houlihan, P.R, Matamala, E, Cabezas-Bratesco, D, Lee, G.Y, Cristofori-Armstrong, B, Dilan, T.L, Sanchez-Martinez, S, Matthies, D, Yan, R, Yu, Z, Ren, D, Brauchi, S.E, Clapham, D.E. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Elife, 12, 2023
|
|
8OR1
 
 | | Co-crystal strucutre of PD-L1 with low molecular weight inhibitor | | Descriptor: | 5-[[5-[[2-chloranyl-3-(2-fluorophenyl)phenyl]methoxy]-2-[(~{E})-2-hydroxyethyliminomethyl]phenoxy]methyl]pyridine-3-carbonitrile, Programmed cell death 1 ligand 1 | | Authors: | Zhang, H, Zhou, S, Wu, C, Zhu, M, Yu, Q, Wang, X, Awadasseid, A, Plewka, J, Magiera-Mularz, K, Wu, Y, Zhang, W. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Design, Synthesis, and Antitumor Activity Evaluation of 2-Arylmethoxy-4-(2,2'-dihalogen-substituted biphenyl-3-ylmethoxy) Benzylamine Derivatives as Potent PD-1/PD-L1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
6ZQN
 
 | | bovine ATP synthase monomer state 3 (combined) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-07-10 | | Release date: | 2020-09-09 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8H5S
 
 | | Crystal structure of Rv3400 from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Beta-phosphoglucomutase, CHLORIDE ION, ... | | Authors: | Singh, L, Karthikeyan, S, Thakur, K.G. | | Deposit date: | 2022-10-13 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural characterization reveals Rv3400 codes for beta-phosphoglucomutase in Mycobacterium tuberculosis.
Protein Sci., 33, 2024
|
|
4Y1V
 
 | | Complex of human Galectin-1 and Galbeta1-3GlcNAc | | Descriptor: | Galectin-1, beta-D-galactopyranose-(1-3)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
5OFJ
 
 | | Crystal structure of N-terminal domain of bifunctional CbXyn10C | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Glycoside hydrolase family 48 | | Authors: | Hakulinen, N, Penttinen, L, Rouvinen, J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Insights into the roles of non-catalytic residues in the active site of a GH10 xylanase with activity on cellulose.
J. Biol. Chem., 292, 2017
|
|
1FMI
 
 | | CRYSTAL STRUCTURE OF HUMAN CLASS I ALPHA1,2-MANNOSIDASE | | Descriptor: | CALCIUM ION, ENDOPLASMIC RETICULUM ALPHA-MANNOSIDASE I, SULFATE ION | | Authors: | Vallee, F, Karaveg, K, Herscovics, A, Moremen, K.W, Howell, P.L. | | Deposit date: | 2000-08-17 | | Release date: | 2001-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for catalysis and inhibition of N-glycan processing class I alpha 1,2-mannosidases.
J.Biol.Chem., 275, 2000
|
|
6PRE
 
 | | SBP RafE in complex with verbascose | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter sugar-binding protein, alpha-D-galactopyranose-(1-6)-alpha-D-galactopyranose-(1-6)-alpha-D-galactopyranose-(1-6)-[beta-D-fructofuranose-(2-1)]alpha-D-glucopyranose | | Authors: | Meier, E.P.W, Boraston, A.B. | | Deposit date: | 2019-07-10 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular analysis of an enigmaticStreptococcus pneumoniaevirulence factor: The raffinose-family oligosaccharide utilization system.
J.Biol.Chem., 294, 2019
|
|
8BY4
 
 | |
8R11
 
 | | Structure of compound 7 bound to SARS-CoV-2 main protease | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2~{S})-2-(3-chlorophenyl)pyrrolidin-1-yl]-2-(5-methylpyridin-3-yl)ethanone, 3C-like proteinase, ... | | Authors: | Mac Sweeney, A, Hazemann, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches.
Rsc Med Chem, 15, 2024
|
|
6UGV
 
 | | Crystal structure of the Fab fragment of anti-TNFa antibody infliximab (Remicade) in a I-centered orthorhombic crystal form, Lot C | | Descriptor: | 1,2-ETHANEDIOL, Infliximab Fab Heavy Chain, Infliximab Fab Light Chain, ... | | Authors: | Lerch, T.F, Sharpe, P, Mayclin, S.J, Edwards, T.E, Polleck, S, Rouse, J.C, Conlan, H.D. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
BioDrugs, 34, 2020
|
|
8QMF
 
 | | Transketolase from Vibrio vulnificus in complex with thiamin pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Ballut, L, Georges, R.N, Octobre, G, Charmantray, F, Doumeche, B. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural determination and kinetic analysis of the transketolase from Vibrio vulnificus reveal unexpected cooperative behavior.
Protein Sci., 33, 2024
|
|
8K3X
 
 | | S. cerevisiae Chs1 in complex with Nikkomycin Z | | Descriptor: | (2S)-{[(2S,3S,4S)-2-amino-4-hydroxy-4-(5-hydroxypyridin-2-yl)-3-methylbutanoyl]amino}[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]acetic acid (non-preferred name), Chitin synthase 1 | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase.
Nat Commun, 14, 2023
|
|
8GEQ
 
 | | E. eligens beta-glucuronidase bound to ceritinib-glucuronide | | Descriptor: | 4-amino-5-chloro-2-{4-(1-beta-D-glucopyranuronosylpiperidin-4-yl)-5-methyl-2-[(propan-2-yl)oxy]anilino}pyrimidine, Beta-glucuronidase | | Authors: | Simpson, J.B, Kowalewski, M.K, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
