9BE2
 
 | |
9BDX
 
 | |
9BDW
 
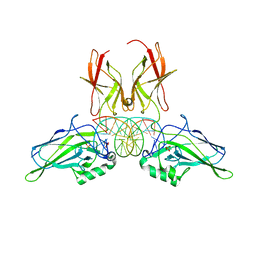
 | | NF-kappaB RelA homo-dimer bound to GC-centric kappaB DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*GP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), DNA (5'-D(P*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*CP*TP*TP*CP*CP*CP*AP*GP*T)-3'), SULFATE ION, ... | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Huang, D, Ghosh, G. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Transient interactions modulate affinity of NF-kappaB transcription factors for DNA
To Be Published
|
|
9BDV
 
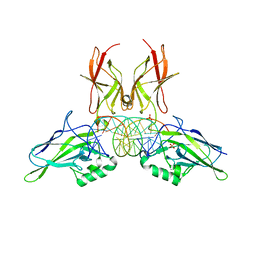
 | | NF-kappaB RelA homo-dimer bound to TA-centric kappaB DNA | | Descriptor: | DNA (5'-D(*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), SULFATE ION, ... | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transient interactions modulate affinity of NF-kappaB transcription factors for DNA
To Be Published
|
|
9BDU
 
 | | NF-kappaB RelA homo-dimer bound to AT-centric kappaB DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), DNA (5'-D(P*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*TP*TP*TP*CP*CP*CP*AP*GP*T)-3'), SULFATE ION, ... | | Authors: | Biswas, T, Huang, D, Ghosh, G. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Transient interactions modulate affinity of NF-kappaB transcription factors for DNA
To Be Published
|
|
9BD3
 
 | | Structure of the MAGEA4 MHD-RAD18 R6BD Complex | | Descriptor: | E3 ubiquitin-protein ligase RAD18, Melanoma antigen A 4 | | Authors: | Forker, K, Zhou, P. | | Deposit date: | 2024-04-10 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal Structure of MAGEA4 MHD-RAD18 R6BD Reveals a Flipped Binding Mode from AlphaFold2 Prediction
Embo J., 2024
|
|
9BD0
 
 | |
9BCZ
 
 | | Chicken 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 (PLCZ1) in complex with calcium and phosphorylated threonine | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Edwards, M.M, Dong, A, Theo-Emegano, N, Seitova, A, Loppnau, P, Leung, R, Li, H, Ilyassov, O, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-10 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Chicken 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 (PLCZ1) in complex with calcium and phosphorylated threonine
To be published
|
|
9BCB
 
 | |
9BBM
 
 | |
9BBL
 
 | |
9BB7
 
 | |
9BB6
 
 | |
9BB5
 
 | |
9BB4
 
 | |
9BB3
 
 | |
9BB2
 
 | |
9BB1
 
 | |
9B94
 
 | |
9B93
 
 | |
9B92
 
 | |
9B91
 
 | |
9B90
 
 | |
9B8Z
 
 | |
9B8Y
 
 | |
