7XL4
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (closed lobe) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7XL3
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (open lobe) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7X76
 
 | |
6BYU
 
 | | X-ray crystal structure of Escherichia coli RNA polymerase (RpoB-H526Y) and ppApp complex | | Descriptor: | (5R)-5-(6-amino-9H-purin-9-yl)-2-({[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}methyl)-4-oxo-4,5-dihydrofuran-3-yl trihydrogen diphosphate, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Murakami, K.S, Molodtsov, V. | | Deposit date: | 2017-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure-function comparisons of (p)ppApp vs (p)ppGpp for Escherichia coli RNA polymerase binding sites and for rrnB P1 promoter regulatory responses in vitro.
Biochim. Biophys. Acta, 1861, 2018
|
|
6C04
 
 | | Mtb RNAP Holo/RbpA/double fork DNA -closed clamp | | Descriptor: | DNA (26-MER), DNA (31-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J, Lilic, M. | | Deposit date: | 2017-12-27 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Fidaxomicin jamsMycobacterium tuberculosisRNA polymerase motions needed for initiation via RbpA contacts.
Elife, 7, 2018
|
|
7X74
 
 | |
7X75
 
 | |
6CCE
 
 | | Crystal structure of a Mycobacterium smegmatis RNA polymerase transcription initiation complex with inhibitor Kanglemycin A | | Descriptor: | 1,2-ETHANEDIOL, DNA (57-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Rifamycin congeners kanglemycins are active against rifampicin-resistant bacteria via a distinct mechanism.
Nat Commun, 9, 2018
|
|
6CCV
 
 | | Crystal structure of a Mycobacterium smegmatis RNA polymerase transcription initiation complex with inhibitor Rifampicin | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Rifamycin congeners kanglemycins are active against rifampicin-resistant bacteria via a distinct mechanism.
Nat Commun, 9, 2018
|
|
6BZO
 
 | | Mtb RNAP Holo/RbpA/Fidaxomicin/upstream fork DNA | | Descriptor: | DNA (26-MER), DNA (32-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2017-12-25 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Fidaxomicin jamsMycobacterium tuberculosisRNA polymerase motions needed for initiation via RbpA contacts.
Elife, 7, 2018
|
|
6C06
 
 | | Mycobacterium tuberculosis RNAP Holo/RbpA/Fidaxomicin | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J, Lilic, M. | | Deposit date: | 2017-12-27 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.15 Å) | | Cite: | Fidaxomicin jamsMycobacterium tuberculosisRNA polymerase motions needed for initiation via RbpA contacts.
Elife, 7, 2018
|
|
6C9Y
 
 | | Cryo-EM structure of E. coli RNAP sigma70 holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Narayanan, A, Vago, F, Li, K, Qayyum, M.Z, Yenool, D, Jiang, W, Murakami, K.S. | | Deposit date: | 2018-01-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Cryo-EM structure ofEscherichia colisigma70RNA polymerase and promoter DNA complex revealed a role of sigma non-conserved region during the open complex formation.
J. Biol. Chem., 293, 2018
|
|
6C05
 
 | | Mycobacterium tuberculosis RNAP Holo/RbpA in relaxed state | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J, Lilic, M. | | Deposit date: | 2017-12-27 | | Release date: | 2018-03-28 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (5.15 Å) | | Cite: | Fidaxomicin jamsMycobacterium tuberculosisRNA polymerase motions needed for initiation via RbpA contacts.
Elife, 7, 2018
|
|
4XLS
 
 | |
4XLR
 
 | |
4XLN
 
 | |
4XLQ
 
 | |
4XSZ
 
 | | Crystal structure of CBR 9393 bound to Escherichia coli RNA polymerase holoenzyme | | Descriptor: | 4-[3-(4-fluorophenyl)-1H-pyrazol-4-yl]-N-[2-(piperazin-1-yl)ethyl]-2-(trifluoromethyl)aniline, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2015-01-22 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.683 Å) | | Cite: | CBR antimicrobials inhibit RNA polymerase via at least two bridge-helix cap-mediated effects on nucleotide addition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XSX
 
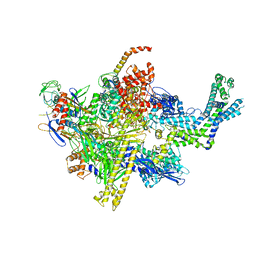 | | Crystal structure of CBR 703 bound to Escherichia coli RNA polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2015-01-22 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.708 Å) | | Cite: | CBR antimicrobials inhibit RNA polymerase via at least two bridge-helix cap-mediated effects on nucleotide addition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XLP
 
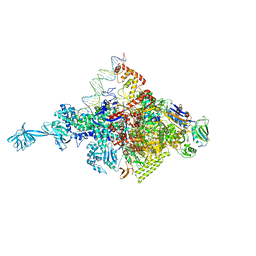 | |
4XSY
 
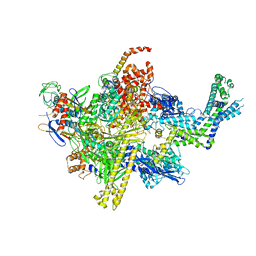 | | Crystal structure of CBR 9379 bound to Escherichia coli RNA polymerase holoenzyme | | Descriptor: | 3-{[(2,6-dichlorophenyl)carbamoyl]amino}-N-hydroxy-N'-phenyl-5-(trifluoromethyl)benzenecarboximidamide, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2015-01-22 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.007 Å) | | Cite: | CBR antimicrobials inhibit RNA polymerase via at least two bridge-helix cap-mediated effects on nucleotide addition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YLP
 
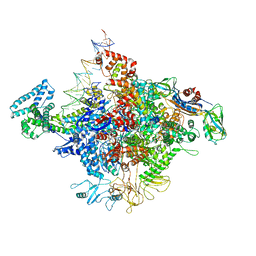 | | E. coli Transcription Initiation Complex - 16-bp spacer and 5-nt RNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuo, Y, Steitz, T.A. | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Crystal Structures of the E. coli Transcription Initiation Complexes with a Complete Bubble.
Mol.Cell, 58, 2015
|
|
4YG2
 
 | | X-ray crystal structur of Escherichia coli RNA polymerase sigma70 holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | X-ray crystal structure of Escherichia coli RNA polymerase sigma70 holoenzyme
J. Biol. Chem., 288, 2013
|
|
4YLO
 
 | | E. coli Transcription Initiation Complex - 16-bp spacer and 4-nt RNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuo, Y, Steitz, T.A. | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Crystal Structures of the E. coli Transcription Initiation Complexes with a Complete Bubble.
Mol.Cell, 58, 2015
|
|
4YLN
 
 | | E. coli Transcription Initiation Complex - 17-bp spacer and 4-nt RNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuo, Y, Steitz, T.A. | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Crystal Structures of the E. coli Transcription Initiation Complexes with a Complete Bubble.
Mol.Cell, 58, 2015
|
|
