1SYR
 
 | |
1SYS
 
 | | Crystal structure of HLA, B*4403, and peptide EEPTVIKKY | | Descriptor: | Beta-2-microglobulin, Sorting nexin 5, leukocyte antigen (HLA) class I molecule | | Authors: | Zernich, D, Purcell, A.W, Macdonald, W.A, Kjer-Nielsen, L, Ely, L.K, Laham, N, Crockford, T, Mifsud, N.A, Tait, B.D, Holdsworth, R, Brooks, A.G, Bottomley, S.P, Beddoe, T, Peh, C.A, Rossjohn, J, McCluskey, J. | | Deposit date: | 2004-04-01 | | Release date: | 2004-10-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion
J.Exp.Med., 200, 2004
|
|
1SYT
 
 | | Crystal structure of signalling protein from goat SPG-40 in the presense of N,N',N''-triacetyl-chitotriose at 2.6A resolution | | Descriptor: | BP40, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kumar, J, Prem Kumar, R, Srivastava, D.B, Sharma, S, Singh, T.P. | | Deposit date: | 2004-04-02 | | Release date: | 2004-04-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of signalling protein from goat SPG-40 in the presense of N,N',N''-triacetyl-chitotriose at 2.6A resolution
to be published
|
|
1SYV
 
 | | HLA-B*4405 complexed to the dominant self ligand EEFGRAYGF | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, major histocompatibility complex, ... | | Authors: | Zernich, D, Purcell, A.W, Macdonald, W.A, Kjer-Nielsen, L, Ely, L.K, Laham, N, Crockford, T, Mifsud, N.A, Tait, B.D, Holdsworth, R, Brooks, A.G, Bottomley, S.P, Beddoe, T, Peh, C.A, Rossjohn, J, McCluskey, J. | | Deposit date: | 2004-04-02 | | Release date: | 2004-10-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion
J.Exp.Med., 200, 2004
|
|
1SYX
 
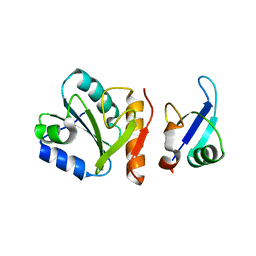 | | The crystal structure of a binary U5 snRNP complex | | Descriptor: | CD2 antigen cytoplasmic tail-binding protein 2, Spliceosomal U5 snRNP-specific 15 kDa protein | | Authors: | Nielsen, T.K, Liu, S, Luhrmann, R, Ficner, R. | | Deposit date: | 2004-04-02 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.345 Å) | | Cite: | Structural basis for the bifunctionality of the U5 snRNP 52K protein (CD2BP2).
J.Mol.Biol., 369, 2007
|
|
1SYY
 
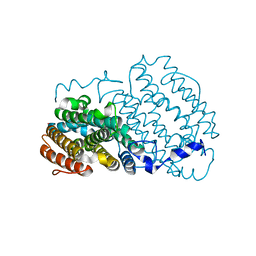 | | Crystal structure of the R2 subunit of ribonucleotide reductase from Chlamydia trachomatis | | Descriptor: | FE (III) ION, LEAD (II) ION, Ribonucleoside-diphosphate reductase beta chain | | Authors: | Hogbom, M, Stenmark, P, Voevodskaya, N, McClarty, G, Graslund, A, Nordlund, P. | | Deposit date: | 2004-04-02 | | Release date: | 2004-07-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The radical site in chlamydial ribonucleotide reductase defines a new R2 subclass.
Science, 305, 2004
|
|
1SYZ
 
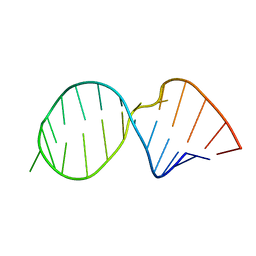 | |
1SZ0
 
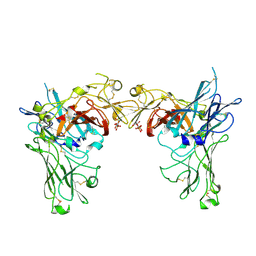 | | N-terminal 3 domains of CI-MPR bound to mannose 6-phosphate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Olson, L.J, Dahms, N.M, Kim, J.-J.P. | | Deposit date: | 2004-04-01 | | Release date: | 2004-06-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The N-terminal carbohydrate recognition site of the cation-independent mannose 6-phosphate receptor
J.Biol.Chem., 279, 2004
|
|
1SZ1
 
 | |
1SZ2
 
 | | Crystal structure of E. coli glucokinase in complex with glucose | | Descriptor: | Glucokinase, beta-D-glucopyranose | | Authors: | Lunin, V.V, Li, Y, Schrag, J.D, Iannuzzi, P, Matte, A, Cygler, M. | | Deposit date: | 2004-04-02 | | Release date: | 2004-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Escherichia coli ATP-dependent glucokinase and its complex with glucose
J.Bacteriol., 186, 2004
|
|
1SZ3
 
 | | CRYSTAL STRUCTURE OF NUDIX HYDROLASE DR1025 IN COMPLEXED WITH GNP AND MG+2 | | Descriptor: | MAGNESIUM ION, MutT/nudix family protein, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Ranatunga, W, Hill, E.E, Mooster, J.L, Holbrook, E.L, Schulze-Gahmen, U, Xu, W, Bessman, M.J, Brenner, S.E, Holbrook, S.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-04-02 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Studies of the Nudix Hydrolase DR1025 From Deinococcus radiodurans and its Ligand Complexes.
J.Mol.Biol., 339, 2004
|
|
1SZ6
 
 | | MISTLETOE LECTIN I FROM VISCUM ALBUM. CRYSTAL STRUCTURE AT 2.05 A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AZIDE ION, BETA-GALACTOSIDE SPECIFIC LECTIN I B CHAIN, ... | | Authors: | Gabdoulkhakov, A.G, Guhlistova, N.E, Lyashenko, A.V, Krauspenhaar, R, Stoeva, S, Voelter, W, Nikonov, S.V, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2004-04-04 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Viscum album Mistletoe Lectin I in native state at 2.05 A resolution, comparison of structure active site conformation in ricin and in viscumin
TO BE PUBLISHED
|
|
1SZ7
 
 | | Crystal structure of Human Bet3 | | Descriptor: | PALMITIC ACID, Trafficking protein particle complex subunit 3 | | Authors: | Turnbull, A.P, Prinz, B, Holz, C, Behlke, J, Schultchen, J, Delbrueck, H, Niesen, F.H, Lang, C, Heinemann, U. | | Deposit date: | 2004-04-05 | | Release date: | 2005-01-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of palmitoylated BET3: insights into TRAPP complex assembly and membrane localization
Embo J., 24, 2005
|
|
1SZ8
 
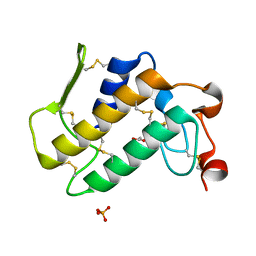 | | Crystal Structure of an Acidic Phospholipase A2 from Naja Naja Sagittifera at 1.5 A resolution | | Descriptor: | ACETIC ACID, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Singh, R.K, Sharma, S, Jabeen, T, Kaur, P, Singh, T.P. | | Deposit date: | 2004-04-05 | | Release date: | 2004-04-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of an Acidic Phospholipase A2 from Naja Naja Sagittifera at 1.5 A Resolution
To be Published
|
|
1SZ9
 
 | |
1SZA
 
 | |
1SZB
 
 | | Crystal structure of the human MBL-associated protein 19 (MAp19) | | Descriptor: | CALCIUM ION, mannose binding lectin-associated serine protease-2 related protein, MAp19 (19kDa) | | Authors: | Gregory, L.A, Thielens, N.M, Arlaud, G.J, Fontecilla-Camps, J.C, Gaboriaud, C. | | Deposit date: | 2004-04-05 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The X-ray structure of human MBL-associated protein 19 (MAp19) and its interaction site with mannan-binding lectin and L-ficolin
J.Biol.Chem., 279, 2004
|
|
1SZC
 
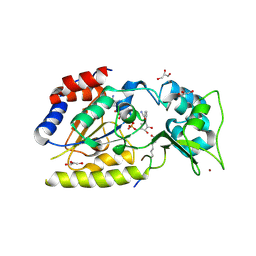 | | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD+-dependent Sir2 histone/protein deacetylases | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhao, K, Harshaw, R, Chai, X, Marmorstein, R. | | Deposit date: | 2004-04-05 | | Release date: | 2004-06-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1SZD
 
 | | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD+-dependent Sir2 histone/protein deacetylases | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhao, K, Harshaw, R, Chai, X, Marmorstein, R. | | Deposit date: | 2004-04-05 | | Release date: | 2004-06-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1SZE
 
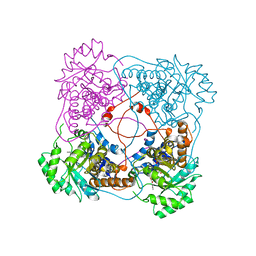 | | L230A mutant flavocytochrome b2 with benzoylformate | | Descriptor: | BENZOYL-FORMIC ACID, Cytochrome b2, mitochondrial, ... | | Authors: | Mowat, C.G, Wehenkel, A, Green, A.J, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2004-04-05 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Altered Substrate Specificity in Flavocytochrome b(2): Structural Insights into the Mechanism of l-Lactate Dehydrogenation
Biochemistry, 43, 2004
|
|
1SZF
 
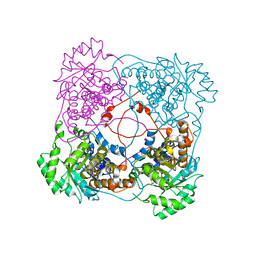 | | A198G:L230A mutant flavocytochrome b2 with pyruvate bound | | Descriptor: | Cytochrome b2, mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Mowat, C.G, Wehenkel, A, Green, A.J, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2004-04-05 | | Release date: | 2004-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Altered Substrate Specificity in Flavocytochrome b(2): Structural Insights into the Mechanism of l-Lactate Dehydrogenation
Biochemistry, 43, 2004
|
|
1SZG
 
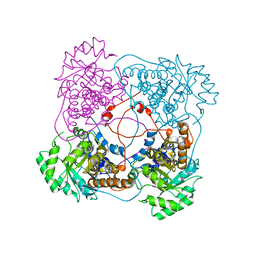 | | A198G:L230A flavocytochrome b2 with sulfite bound | | Descriptor: | Cytochrome b2, mitochondrial, N-SULFO-FLAVIN MONONUCLEOTIDE | | Authors: | Mowat, C.G, Wehenkel, A, Green, A.J, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2004-04-05 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Altered Substrate Specificity in Flavocytochrome b(2): Structural Insights into the Mechanism of l-Lactate Dehydrogenation
Biochemistry, 43, 2004
|
|
1SZH
 
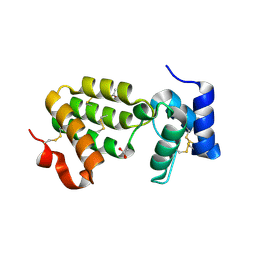 | | Crystal Structure of C. elegans HER-1 | | Descriptor: | ACETATE ION, Her-1 protein | | Authors: | Hamaoka, B.Y, Dann III, C.E, Geisbrecht, B.V, Leahy, D.J. | | Deposit date: | 2004-04-05 | | Release date: | 2004-08-10 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Caenorhabditis elegans HER-1 and characterization of the interaction between HER-1 and TRA-2A.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1SZI
 
 | |
1SZJ
 
 | |
