1J8I
 
 | | Solution Structure of Human Lymphotactin | | Descriptor: | Lymphotactin | | Authors: | Kuloglu, E.S, McCaslin, D.R, Markley, J.L, Pauza, C.D, Volkman, B.F. | | Deposit date: | 2001-05-21 | | Release date: | 2001-10-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Monomeric solution structure of the prototypical 'C' chemokine lymphotactin.
Biochemistry, 40, 2001
|
|
1IVS
 
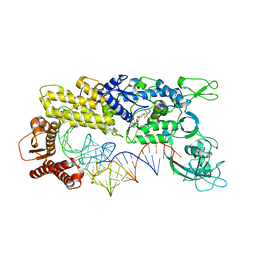 | | CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS VALYL-TRNA SYNTHETASE COMPLEXED WITH TRNA(VAL) AND VALYL-ADENYLATE ANALOGUE | | Descriptor: | N-[VALINYL]-N'-[ADENOSYL]-DIAMINOSUFONE, Valyl-tRNA synthetase, tRNA (Val) | | Authors: | Fukai, S, Nureki, O, Sekine, S.-I, Shimada, A, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-29 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of molecular interactions for tRNA(Val) recognition by valyl-tRNA synthetase
RNA, 9, 2003
|
|
8CH7
 
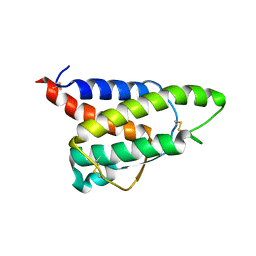 | | RDC-refined Interleukin-4 (wild type) pH 5.6 | | Descriptor: | Interleukin-4 | | Authors: | Vaz, D.C, Rodrigues, J.R, Loureiro-Ferreira, N, Mueller, T, Sebald, W, Redfield, C, Brito, R.M.M. | | Deposit date: | 2023-02-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Lessons on protein structure from interleukin-4: All disulfides are not created equal.
Proteins, 92, 2024
|
|
8CGF
 
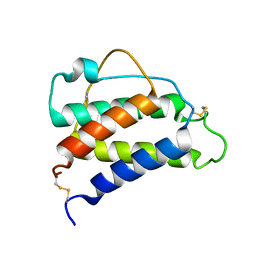 | | Interleukin-4 (wild type) pH 2.4 | | Descriptor: | Interleukin-4 | | Authors: | Vaz, D.C, Rodrigues, J.R, Loureiro-Ferreira, N, Mueller, T, Sebald, W, Redfield, C, Brito, R.M.M. | | Deposit date: | 2023-02-04 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Lessons on protein structure from interleukin-4: All disulfides are not created equal.
Proteins, 92, 2024
|
|
7T1Z
 
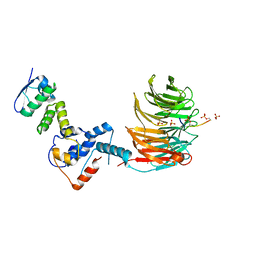 | | Structure of the Fbw7-Skp1-MycNdegron complex | | Descriptor: | F-box/WD repeat-containing protein 7, Myc proto-oncogene N terminal degron, S-phase kinase-associated protein 1, ... | | Authors: | Wang, B, Rusnac, D.V, Clurman, B.E, Zheng, N. | | Deposit date: | 2021-12-02 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Two diphosphorylated degrons control c-Myc degradation by the Fbw7 tumor suppressor.
Sci Adv, 8, 2022
|
|
1J3U
 
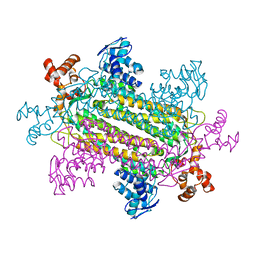 | |
1ITM
 
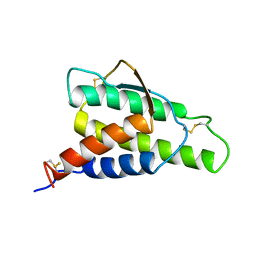 | | ANALYSIS OF THE SOLUTION STRUCTURE OF HUMAN INTERLEUKIN 4 DETERMINED BY HETERONUCLEAR THREE-DIMENSIONAL NUCLEAR MAGNETIC RESONANCE TECHNIQUES | | Descriptor: | INTERLEUKIN-4 | | Authors: | Redfield, C, Smith, L.J, Boyd, J, Lawrence, G.M.P, Edwards, R.G, Gershater, C.J, Smith, R.A.G, Dobson, C.M. | | Deposit date: | 1994-02-28 | | Release date: | 1994-05-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Analysis of the solution structure of human interleukin-4 determined by heteronuclear three-dimensional nuclear magnetic resonance techniques.
J.Mol.Biol., 238, 1994
|
|
1IR3
 
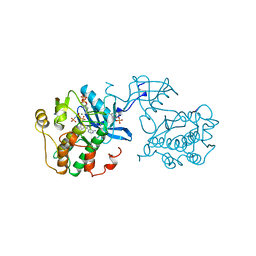 | |
7T6F
 
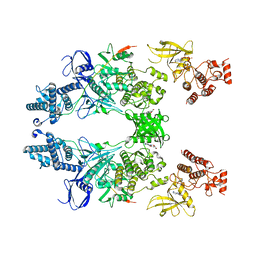 | | Structure of active Janus Kinase (JAK) dimer complexed with cytokine receptor intracellular domain | | Descriptor: | ADENOSINE, ADENOSINE-5'-DIPHOSPHATE, Interferon lambda receptor 1, ... | | Authors: | Glassman, C.R, Tsutsumi, N, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-12-13 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a Janus kinase cytokine receptor complex reveals the basis for dimeric activation.
Science, 376, 2022
|
|
1IJ2
 
 | |
6PXW
 
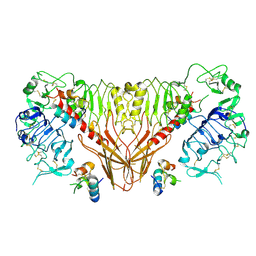 | | Cryo-EM structure of full-length insulin receptor bound to 4 insulin. 3D refinement was focused on the top part of the receptor complex. | | Descriptor: | Insulin, Insulin receptor | | Authors: | Uchikawa, E, Choi, E, Shang, G.J, Yu, H.T, Bai, X.C. | | Deposit date: | 2019-07-28 | | Release date: | 2019-09-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex.
Elife, 8, 2019
|
|
1INR
 
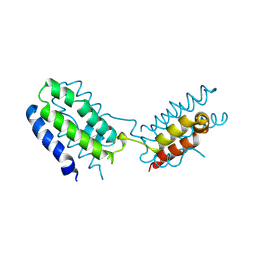 | | CYTOKINE SYNTHESIS | | Descriptor: | INTERLEUKIN-10 | | Authors: | Walter, M.R. | | Deposit date: | 1995-07-31 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of interleukin 10 reveals an interferon gamma-like fold.
Biochemistry, 34, 1995
|
|
1IZE
 
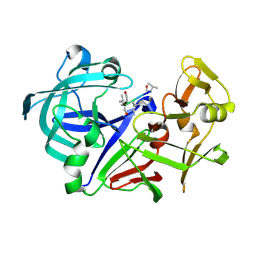 | | Crystal structure of Aspergillus oryzae Aspartic proteinase complexed with pepstatin | | Descriptor: | Pepstatin, alpha-D-mannopyranose, aspartic proteinase | | Authors: | Kamitori, S, Ohtaki, A, Ino, H, Takeuchi, M. | | Deposit date: | 2002-10-02 | | Release date: | 2003-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Aspergillus oryzae aspartic proteinase and its complex with an inhibitor pepstatin at 1.9A resolution.
J.Mol.Biol., 326, 2003
|
|
7SQC
 
 | | Ciliary C1 central pair apparatus isolated from Chlamydomonas reinhardtii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CPC1, Calmodulin, ... | | Authors: | Gui, M, Wang, X, Dutcher, S.K, Brown, A, Zhang, R. | | Deposit date: | 2021-11-05 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ciliary central apparatus structure reveals mechanisms of microtubule patterning.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1J1C
 
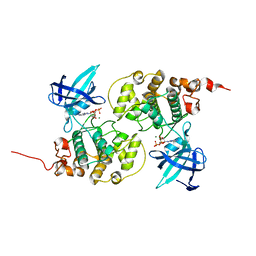 | | Binary complex structure of human tau protein kinase I with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glycogen synthase kinase-3 beta, MAGNESIUM ION | | Authors: | Aoki, M, Yokota, T, Sugiura, I, Sasaki, C, Hasegawa, T, Okumura, C, Kohno, T, Sugio, S, Matsuzaki, T. | | Deposit date: | 2002-12-03 | | Release date: | 2003-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into nucleotide recognition in tau-protein kinase I/glycogen synthase kinase 3 beta.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6Q0T
 
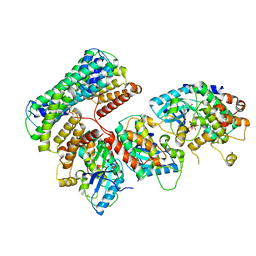 | | Structure of a MAPK pathway complex | | Descriptor: | 14-3-3 protein zeta, 5-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)imidazo[1,5-a]pyridine-6-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Park, E, Rawson, S, Jeon, H, Eck, M.J. | | Deposit date: | 2019-08-02 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Architecture of autoinhibited and active BRAF-MEK1-14-3-3 complexes.
Nature, 575, 2019
|
|
1J7V
 
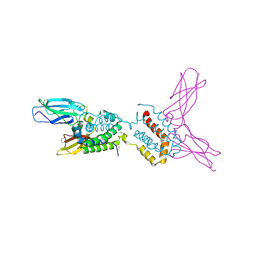 | | HUMAN IL-10 / IL-10R1 COMPLEX | | Descriptor: | INTERLEUKIN-10, INTERLEUKIN-10 RECEPTOR ALPHA CHAIN | | Authors: | Josephson, K, Logsdon, N, Walter, M.R. | | Deposit date: | 2001-05-18 | | Release date: | 2001-09-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the IL-10/IL-10R1 complex reveals a shared receptor binding site.
Immunity, 15, 2001
|
|
1IJ0
 
 | |
1Q07
 
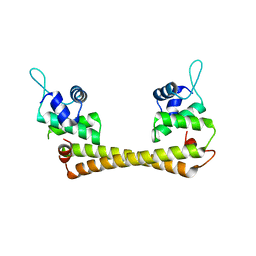 | | Crystal structure of the Au(I) form of E. coli CueR, a copper efflux regulator | | Descriptor: | GOLD ION, Transcriptional regulator cueR | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
4OWI
 
 | | peptide structure | | Descriptor: | p53LZ2 | | Authors: | Lee, J.-H. | | Deposit date: | 2014-02-02 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.202 Å) | | Cite: | Protein grafting of p53TAD onto a leucine zipper scaffold generates a potent HDM dual inhibitor.
Nat Commun, 5, 2014
|
|
1IJ1
 
 | |
1J2B
 
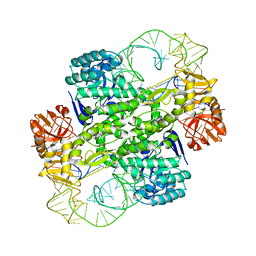 | | Crystal Structure Of Archaeosine tRNA-Guanine Transglycosylase Complexed With lambda-form tRNA(Val) | | Descriptor: | Archaeosine tRNA-guanine transglycosylase, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ishitani, R, Nureki, O, Nameki, N, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-29 | | Release date: | 2003-05-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Alternative Tertiary Structure of tRNA for Recognition by a Posttranscriptional Modification Enzyme
Cell(Cambridge,Mass.), 113, 2003
|
|
1J5E
 
 | | Structure of the Thermus thermophilus 30S Ribosomal Subunit | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Wimberly, B.T, Brodersen, D.E, Clemons Jr, W.M, Morgan-Warren, R, Carter, A.P, Vonrhein, C, Hartsch, T, Ramakrishnan, V. | | Deposit date: | 2002-04-08 | | Release date: | 2002-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of the 30S ribosomal subunit.
Nature, 407, 2000
|
|
4OHT
 
 | | Crystal structure of succinic semialdehyde dehydrogenase from Streptococcus pyogenes in complex with NADP+ as the cofactor | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Succinate-semialdehyde dehydrogenase | | Authors: | Park, S.A, Jang, E.H, Chi, Y.M, Lee, K.S. | | Deposit date: | 2014-01-18 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and Structural Characterization for Cofactor Preference of Succinic Semialdehyde Dehydrogenase from Streptococcus pyogenes.
Mol.Cells, 37, 2014
|
|
1IZD
 
 | | Crystal structure of Aspergillus oryzae Aspartic Proteinase | | Descriptor: | Aspartic proteinase, alpha-D-mannopyranose | | Authors: | Kamitori, S, Ohtaki, A, Ino, H, Takeuchi, M. | | Deposit date: | 2002-10-02 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Aspergillus oryzae Aspartic Proteinase and its Complex with an Inhibitor Pepstatin at 1.9 A Resolution
J.Mol.Biol., 326, 2003
|
|
