7ME9
 
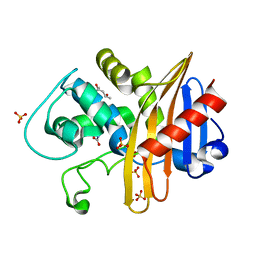 | | CDD-1 beta-lactamase in imidazole/MPD 30 seconds avibactam complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, SULFATE ION | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
4U6G
 
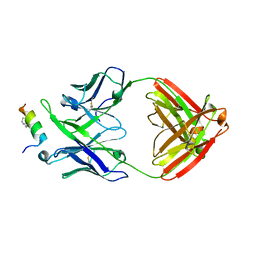 | | Crystal structure of 10E8 Fab in complex with a hydrocarbon-stapled HIV-1 gp41 MPER peptide | | Descriptor: | 10E8 Fab Heavy Chain, 10E8 Fab Light Chain, SAH-MPER(662-683KKK)(B,q) | | Authors: | Ofek, G, Bird, G.H, Walensky, L.D, Kwong, P.D. | | Deposit date: | 2014-07-28 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4.2012 Å) | | Cite: | Structural Fidelity and Neutralizing Antibody Binding Capacity of
Hydrocarbon-Stapled HIV-1 Antigens
To be published
|
|
8VLL
 
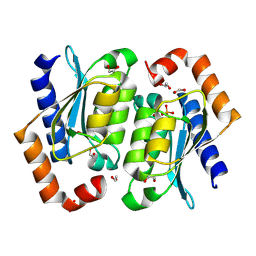 | | Crystal structure of the yeast cytosine deaminase (yCD) M100W mutant | | Descriptor: | 1,2-ETHANEDIOL, Cytosine deaminase, PHOSPHATE ION, ... | | Authors: | Picard, M.-E, Grenier, J, Despres, P.C, Dube, A.K, Landry, C.R, Shi, R. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Compensatory mutations potentiate constructive neutral evolution by gene duplication.
Science, 385, 2024
|
|
6ZSP
 
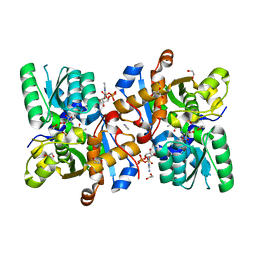 | | Human serine racemase bound to ATP and malonate. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Koulouris, C.R, Bax, B.D, Roe, S.M, Atack, J.R. | | Deposit date: | 2020-07-16 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
6ZT3
 
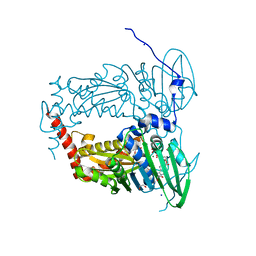 | | N-terminal 47 kDa fragment of the Mycobacterium smegmatis DNA Gyrase B subunit complexed with ADPNP | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Feng, L, Mundy, J.E.A, Stevenson, C.E.M, Mitchenall, L.A, Lawson, D.M, Mi, K, Maxwell, A. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The pentapeptide-repeat protein, MfpA, interacts with mycobacterial DNA gyrase as a DNA T-segment mimic.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ZYF
 
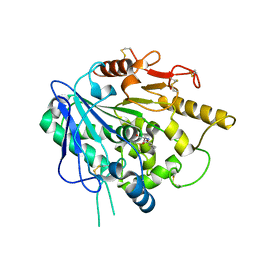 | | Notum_Ghrelin complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-07-31 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Notum deacylates octanoylated ghrelin.
Mol Metab, 49, 2021
|
|
4UZG
 
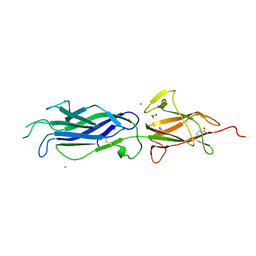 | | Crystal structure of group B streptococcus pilus 2b backbone protein SAK_1440 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SURFACE PROTEIN SPB1 | | Authors: | Malito, E, Cozzi, R, Bottomley, M.J. | | Deposit date: | 2014-09-05 | | Release date: | 2015-05-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structure and assembly of group B streptococcus pilus 2b backbone protein.
PLoS ONE, 10, 2015
|
|
4US5
 
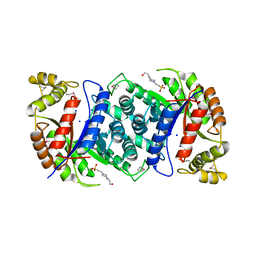 | | Crystal Structure of apo-MsnO8 | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Maier, S, Pflueger, T, Loesgen, S, Asmus, K, Broetz, E, Paululat, T, Zeeck, A, Andrade, S, Bechthold, A. | | Deposit date: | 2014-07-03 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights Into the Bioactivity of Mensacarcin and Epoxide Formation by Msno8.
Chembiochem, 15, 2014
|
|
8RPA
 
 | | Crystal structure of Zea mays adenosine kinase 3 (ZmADK3) in complex with AP5A | | Descriptor: | 1,2-ETHANEDIOL, Adenosine kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ... | | Authors: | Morera, S, Kopecny, D, Vigouroux, A. | | Deposit date: | 2024-01-13 | | Release date: | 2025-01-29 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | A monomer-dimer switch modulates the activity of plant adenosine kinase.
J.Exp.Bot., 76, 2025
|
|
6TVJ
 
 | |
8AKF
 
 | |
1FYV
 
 | | CRYSTAL STRUCTURE OF THE TIR DOMAIN OF HUMAN TLR1 | | Descriptor: | TOLL-LIKE RECEPTOR 1 | | Authors: | Xu, Y, Tao, X, Shen, B, Horng, T, Medzhitov, R, Manley, J.L, Tong, L. | | Deposit date: | 2000-10-03 | | Release date: | 2000-11-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for signal transduction by the Toll/interleukin-1 receptor domains.
Nature, 408, 2000
|
|
8AK9
 
 | |
5VM0
 
 | | The hapten triclocarban bound to the single domain camelid nanobody VHH T9 | | Descriptor: | 1,2-ETHANEDIOL, Camelid Nanobody VHH T9, N-(4-chlorophenyl)-N'-(3,4-dichlorophenyl)urea | | Authors: | Wogulis, L.A, Wogulis, M.D, Tabares-da Rosa, S, Gonzalez-Sapienza, G, Wilson, D.K. | | Deposit date: | 2017-04-26 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure and specificity of several triclocarban-binding single domain camelid antibody fragments.
J. Mol. Recognit., 32, 2019
|
|
7TUX
 
 | | Crystal Structure of Plasmodium falciparum Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase in complex with [(3S)-4-Hydroxy-3-[({2-amino-4-hydroxy-5H-pyrrolo[3,2-d]pyrimidin-7-yl}methyl)amino]butyl]phosphonic acid | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Hypoxanthine-guanine-xanthine phosphoribosyltransferase, ... | | Authors: | Harijan, R.K, Minnow, Y.V.T, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2022-02-03 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Inhibition and Mechanism of Plasmodium falciparum Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase.
Acs Chem.Biol., 17, 2022
|
|
8EX5
 
 | |
5ZO3
 
 | | apo form of the nuclease | | Descriptor: | 1,2-ETHANEDIOL, Putative 3'-5' exonuclease family protein, SULFATE ION | | Authors: | Yuan, Z.L, Gu, L.C. | | Deposit date: | 2018-04-12 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.493 Å) | | Cite: | NrnC, an RNase D-Like Protein FromAgrobacterium, Is a Novel Octameric Nuclease That Specifically Degrades dsDNA but Leaves dsRNA Intact.
Front Microbiol, 9, 2018
|
|
8EX7
 
 | |
8EX8
 
 | |
1G05
 
 | | HETEROCYCLE-BASED MMP INHIBITOR WITH P2'SUBSTITUENTS | | Descriptor: | 1-BENZYL-3-(4-METHOXY-BENZENESULFONYL)-6-OXO-HEXAHYDRO-PYRIMIDINE-4-CARBOXYLIC ACID HYDROXYAMIDE, CALCIUM ION, STROMELYSIN-1 PRECURSOR, ... | | Authors: | Pikul, S, Dunham, K.M, Almstead, N.G, De, B, Natchus, M.G, Taiwo, Y.O, Williams, L.E, Hynd, B.A, Hsieh, L.C, Janusz, M.J. | | Deposit date: | 2000-10-05 | | Release date: | 2001-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Heterocycle-based MMP inhibitors with P2' substituents.
Bioorg.Med.Chem.Lett., 11, 2001
|
|
9FWO
 
 | | Crystal Structure of SARS-CoV-2 NSP10-NSP14 (ExoN) in complex with VT00216 | | Descriptor: | 1-methylpyrrole-2-carboxamide, Guanine-N7 methyltransferase nsp14, MAGNESIUM ION, ... | | Authors: | Krojer, T, Kozielski, F, Sele, C, Nyblom, M, Fisher, S.Z, Knecht, W. | | Deposit date: | 2024-06-30 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.179 Å) | | Cite: | Crystal Structure of SARS-CoV-2 NSP10-NSP14 (ExoN) in complex with VT00216
To Be Published
|
|
9FXV
 
 | |
7EZ0
 
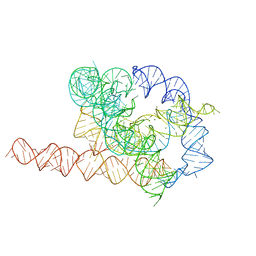 | | Apo L-21 ScaI Tetrahymena ribozyme | | Descriptor: | Apo L-21 ScaI Tetrahymena ribozyme, MAGNESIUM ION | | Authors: | Su, Z, Zhang, K, Kappel, K, Luo, B, Das, R, Chiu, W. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Cryo-EM structures of full-length Tetrahymena ribozyme at 3.1 angstrom resolution.
Nature, 596, 2021
|
|
6W5E
 
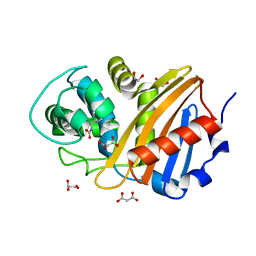 | | Class D beta-lactamase BSU-2 | | Descriptor: | 1,2-ETHANEDIOL, BSU-2 beta-lactamase, MALONATE ION | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A surface loop modulates activity of the Bacillus class D beta-lactamases.
J.Struct.Biol., 211, 2020
|
|
7VTI
 
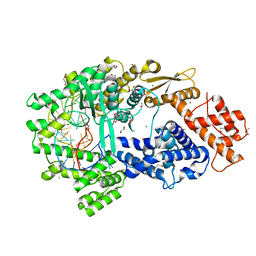 | | Crystal structure of the Cas13bt3-crRNA binary complex | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Nakagawa, R, Takeda, N.S, Tomita, A, Hirano, H, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-10-29 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and engineering of the minimal type VI CRISPR-Cas13bt3.
Mol.Cell, 82, 2022
|
|
