4WB4
 
 | | wt SA11 NSP4_CCD | | Descriptor: | CALCIUM ION, Non-structural glycoprotein NSP4 | | Authors: | Viskovska, M, Sastri, N.P, Hyser, J.M, Tanner, M.R, Horton, L.B, Sankaran, B, Prasad, B.V.V, Estes, M.K. | | Deposit date: | 2014-09-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Plasticity of the Coiled-Coil Domain of Rotavirus NSP4.
J.Virol., 88, 2014
|
|
3S5N
 
 | | Crystal Structure of Human 4-hydroxy-2-oxoglutarate Aldolase | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-2-oxoglutarate aldolase, mitochondrial, ... | | Authors: | Riedel, T.J, Lowther, W.T. | | Deposit date: | 2011-05-23 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Studies of Human 4-hydroxy-2-oxoglutarate Aldolase: Implications for Hydroxyproline Metabolism in Primary Hyperoxaluria.
Plos One, 6, 2011
|
|
3S5S
 
 | |
4WB9
 
 | | Human ALDH1A1 complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Morgan, C.A, Hurley, T.D. | | Deposit date: | 2014-09-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Development of a high-throughput in vitro assay to identify selective inhibitors for human ALDH1A1.
Chem.Biol.Interact., 234, 2015
|
|
3JWD
 
 | | Structure of HIV-1 gp120 with gp41-Interactive Region: Layered Architecture and Basis of Conformational Mobility | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB 48D HEAVY CHAIN, FAB 48D LIGHT CHAIN, ... | | Authors: | Pancera, M, Majeed, S, Ban, Y.A, Chen, L, Huang, C.C, Kong, L, Kwon, Y.D, Stuckey, J, Zhou, T, Robinson, J.E, Schief, W.R, Sodroski, J, Wyatt, R, Kwong, P.D. | | Deposit date: | 2009-09-18 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3SFZ
 
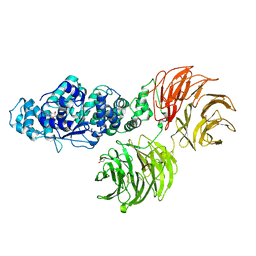 | | Crystal structure of full-length murine Apaf-1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Apoptotic peptidase activating factor 1, GAMMA-BUTYROLACTONE | | Authors: | Eschenburg, S, Reubold, T.F. | | Deposit date: | 2011-06-14 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of full-length Apaf-1: how the death signal is relayed in the mitochondrial pathway of apoptosis.
Structure, 19, 2011
|
|
3SGH
 
 | |
4WCI
 
 | | Crystal structure of the 1st SH3 domain from human CD2AP (CMS) in complex with a proline-rich peptide (aa 378-393) from human RIN3 | | Descriptor: | CD2-associated protein, Ras and Rab interactor 3, SULFATE ION | | Authors: | Rouka, E, Simister, P.C, Janning, M, Kirsch, K.H, Krojer, T, Knapp, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Feller, S.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-04 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Differential Recognition Preferences of the Three Src Homology 3 (SH3) Domains from the Adaptor CD2-associated Protein (CD2AP) and Direct Association with Ras and Rab Interactor 3 (RIN3).
J.Biol.Chem., 290, 2015
|
|
4WCW
 
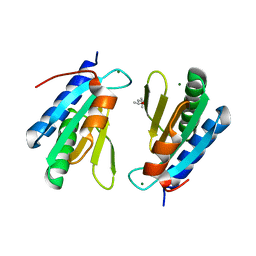 | | Ribosomal silencing factor during starvation or stationary phase (RsfS) from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, Ribosomal silencing factor RsfS | | Authors: | Li, X, Sun, Q, Jiang, C, Yang, K, Hung, L, Zhang, J, Sacchettini, J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Ribosomal Silencing Factor Bound to Mycobacterium tuberculosis Ribosome.
Structure, 23, 2015
|
|
3SHM
 
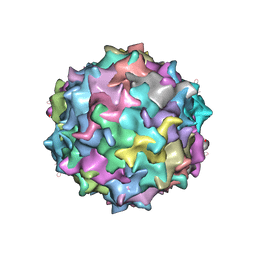 | |
4WD0
 
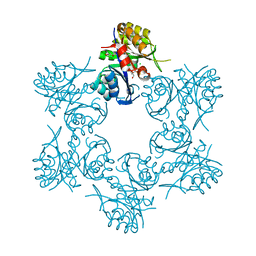 | | Crystal structure of HisAp form Arthrobacter aurescens | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of HisAp form Arthrobacter aurescens
To Be Published
|
|
3S6M
 
 | |
3S6W
 
 | | Crystal structure of Tudor domain of human TDRD3 | | Descriptor: | ISOPROPYL ALCOHOL, Tudor domain-containing protein 3 | | Authors: | Liu, H.P, Xu, R.M. | | Deposit date: | 2011-05-26 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of TDRD3 and methyl-arginine binding characterization of TDRD3, SMN and SPF30
Plos One, 7, 2012
|
|
3S7R
 
 | |
4WD3
 
 | | Crystal structure of an L-amino acid ligase RizA | | Descriptor: | L-amino acid ligase | | Authors: | Kagawa, W, Arai, T, Kino, K, Kurumizaka, H. | | Deposit date: | 2014-09-06 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of RizA, an L-amino-acid ligase from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4WDC
 
 | |
3SM9
 
 | | Crystal Structure of Metabotropic glutamate receptor 3 precursor in presence of LY341495 antagonist | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, CHLORIDE ION, Metabotropic glutamate receptor 3, ... | | Authors: | Wernimont, A.K, Dong, A, Seitova, A, Crombet, L, Khutoreskaya, G, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Cossar, D, Dobrovetsky, E, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-27 | | Release date: | 2011-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structure of Metabotropic glutamate receptor 3 precursor in presence of LY341495 antagonist
TO BE PUBLISHED
|
|
3S93
 
 | | Crystal structure of conserved motif in TDRD5 | | Descriptor: | Tudor domain-containing protein 5, UNKNOWN ATOM OR ION | | Authors: | Chao, X, Tempel, W, Bian, C, Kania, J, Wernimont, A.K, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-31 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of conserved motif in TDRD5
to be published
|
|
4WEK
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 with a R4 substituted vinyl monocarbam | | Descriptor: | (3S,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-3-ethenyl-4-formyl-1-[({3-(5-hydroxy-4-oxo-3,4-dihydropyridin-2-yl)-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-10,10-dimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
4UWB
 
 | | Fibroblast growth factor receptor 1 kinase in complex with JK-P5 | | Descriptor: | 1,2-ETHANEDIOL, FIBROBLAST GROWTH FACTOR RECEPTOR 1, N-[4-(4-methylpiperazin-1-yl)phenyl]-1H-indazole-3-carboxamide, ... | | Authors: | Beeston, H, Tucker, J, Kankanala, J. | | Deposit date: | 2014-08-11 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Validation of IMS-MS as a screening tool to identify type II kinase inhibitors of FGFR1 kinase.
Rapid Commun Mass Spectrom, 2021
|
|
3S9V
 
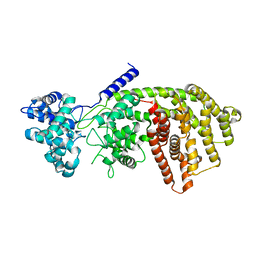 | | abietadiene synthase from Abies grandis | | Descriptor: | Abietadiene synthase, chloroplastic | | Authors: | Zhou, K, Hoy, J.A, Mann, F.M, Honzatko, R.B, Peters, R.J. | | Deposit date: | 2011-06-02 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into diterpene cyclization from structure of bifunctional abietadiene synthase from Abies grandis.
J.Biol.Chem., 287, 2012
|
|
4UWC
 
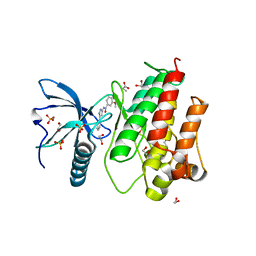 | | Fibroblast growth factor receptor 1 kinase in complex with JK-P3 | | Descriptor: | 1,2-ETHANEDIOL, 3,4-dimethoxy-N-(5-phenyl-1H-pyrazol-3-yl)benzamide, FIBROBLAST GROWTH FACTOR RECEPTOR 1, ... | | Authors: | Beeston, H, Tucker, J, Kankanala, J. | | Deposit date: | 2014-08-11 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Validation of IMS-MS as a screening tool to identify type II kinase inhibitors of FGFR1 kinase.
Rapid Commun Mass Spectrom, 2021
|
|
4UUG
 
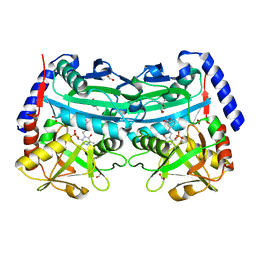 | | The (R)-selective amine transaminase from Aspergillus fumigatus with inhibitor bound | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, ... | | Authors: | Thomsen, M, Hinrichs, W. | | Deposit date: | 2014-07-28 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Characterization of the Dual Substrate Recognition of the (R)-Selective Amine Transaminase from Aspergillus Fumigatus
FEBS J., 282, 2015
|
|
3SN0
 
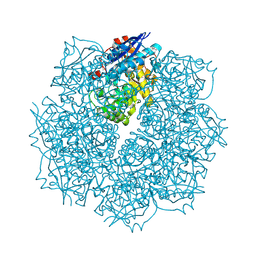 | | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and fumarate | | Descriptor: | CHLORIDE ION, FUMARIC ACID, MAGNESIUM ION, ... | | Authors: | Bonanno, J.B, Patskovsky, Y, Toro, R, Dickey, M, Bain, K.T, Wu, B, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and fumarate
To be Published
|
|
4UU7
 
 | | Crystal structure of zebrafish Sirtuin 5 in complex with 3-methyl- succinylated CPS1-peptide | | Descriptor: | (2R)-2-methylbutanedioic acid, (2S)-2-methylbutanedioic acid, 1,2-ETHANEDIOL, ... | | Authors: | Pannek, M, Gertz, M, Steegborn, C. | | Deposit date: | 2014-07-24 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Chemical Probing of the Human Sirtuin 5 Active Site Reveals its Substrate Acyl Specificity and Peptide-Based Inhibitors.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
