1CY1
 
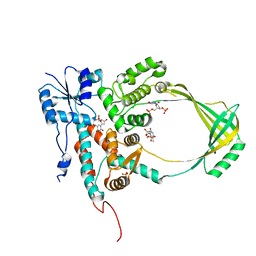 | | COMPLEX OF E.COLI DNA TOPOISOMERASE I WITH 5'PTPTPT | | Descriptor: | DNA TOPOISOMERASE I, PHOSPHATE ION, THYMIDINE-3',5'-DIPHOSPHATE, ... | | Authors: | Feinberg, H, Changela, A, Mondragon, A. | | Deposit date: | 1999-08-31 | | Release date: | 2000-03-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein-nucleotide interactions in E. coli DNA topoisomerase I.
Nat.Struct.Biol., 6, 1999
|
|
1CJ0
 
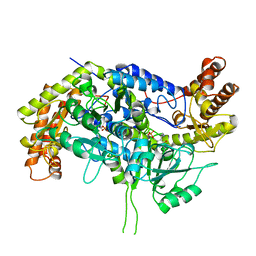 | | CRYSTAL STRUCTURE OF RABBIT CYTOSOLIC SERINE HYDROXYMETHYLTRANSFERASE AT 2.8 ANGSTROM RESOLUTION | | Descriptor: | PROTEIN (SERINE HYDROXYMETHYLTRANSFERASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Scarsdale, J.N, Kazanina, G, Radaev, S, Schirch, V, Wright, H.T. | | Deposit date: | 1999-04-20 | | Release date: | 1999-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of rabbit cytosolic serine hydroxymethyltransferase at 2.8 A resolution: mechanistic implications.
Biochemistry, 38, 1999
|
|
1D40
 
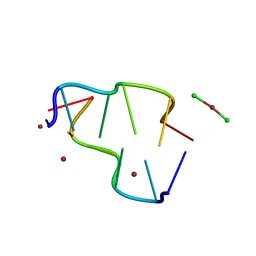 | | BASE SPECIFIC BINDING OF COPPER(II) TO Z-DNA: THE 1.3-ANGSTROMS SINGLE CRYSTAL STRUCTURE OF D(M5CGUAM5CG) IN THE PRESENCE OF CUCL2 | | Descriptor: | COPPER (II) CHLORIDE, COPPER (II) ION, DNA (5'-D(*(5CM)P*(CU)GP*UP*AP*(5CM)P*(CU)G)-3') | | Authors: | Geierstanger, B.H, Kagawa, T.F, Chen, S.-L, Quigley, G.J, Ho, P.S. | | Deposit date: | 1991-05-07 | | Release date: | 1992-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Base-specific binding of copper(II) to Z-DNA. The 1.3-A single crystal structure of d(m5CGUAm5CG) in the presence of CuCl2.
J.Biol.Chem., 266, 1991
|
|
1CR6
 
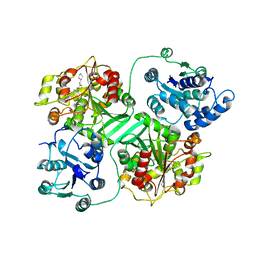 | | CRYSTAL STRUCTURE OF MURINE SOLUBLE EPOXIDE HYDROLASE COMPLEXED WITH CPU INHIBITOR | | Descriptor: | EPOXIDE HYDROLASE, N-CYCLOHEXYL-N'-(PROPYL)PHENYL UREA | | Authors: | Argiriadi, M.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 1999-08-13 | | Release date: | 1999-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Detoxification of environmental mutagens and carcinogens: structure, mechanism, and evolution of liver epoxide hydrolase.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1CTT
 
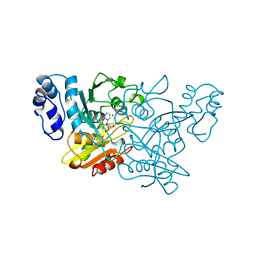 | | TRANSITION-STATE SELECTIVITY FOR A SINGLE OH GROUP DURING CATALYSIS BY CYTIDINE DEAMINASE | | Descriptor: | 3,4-DIHYDRO-1H-PYRIMIDIN-2-ONE NUCLEOSIDE, CYTIDINE DEAMINASE, ZINC ION | | Authors: | Xiang, S, Short, S.A, Wolfenden, R, Carter, C.W. | | Deposit date: | 1995-02-11 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Transition-state selectivity for a single hydroxyl group during catalysis by cytidine deaminase.
Biochemistry, 34, 1995
|
|
1CF7
 
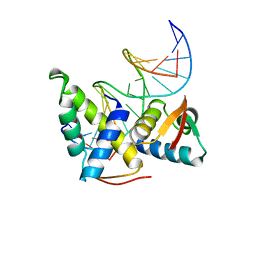 | | STRUCTURAL BASIS OF DNA RECOGNITION BY THE HETERODIMERIC CELL CYCLE TRANSCRIPTION FACTOR E2F-DP | | Descriptor: | DNA (5'-D(*AP*TP*TP*TP*TP*CP*GP*CP*GP*CP*GP*GP*TP*TP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*CP*CP*GP*CP*GP*CP*GP*AP*AP*AP*A)-3'), PROTEIN (TRANSCRIPTION FACTOR DP-2), ... | | Authors: | Zheng, N, Fraenkel, E, Pabo, C.O, Pavletich, N.P. | | Deposit date: | 1999-03-24 | | Release date: | 1999-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP.
Genes Dev., 13, 1999
|
|
1CSV
 
 | |
1CSU
 
 | |
1CSX
 
 | |
1CSW
 
 | |
1CEY
 
 | | ASSIGNMENTS, SECONDARY STRUCTURE, GLOBAL FOLD, AND DYNAMICS OF CHEMOTAXIS Y PROTEIN USING THREE-AND FOUR-DIMENSIONAL HETERONUCLEAR (13C,15N) NMR SPECTROSCOPY | | Descriptor: | CHEY | | Authors: | Moy, F.J, Lowry, D.F, Matsumura, P, Dahlquist, F.W, Krywko, J.E, Domaille, P.J. | | Deposit date: | 1994-11-23 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Assignments, secondary structure, global fold, and dynamics of chemotaxis Y protein using three- and four-dimensional heteronuclear (13C,15N) NMR spectroscopy.
Biochemistry, 33, 1994
|
|
1CFH
 
 | | STRUCTURE OF THE METAL-FREE GAMMA-CARBOXYGLUTAMIC ACID-RICH MEMBRANE BINDING REGION OF FACTOR IX BY TWO-DIMENSIONAL NMR SPECTROSCOPY | | Descriptor: | COAGULATION FACTOR IX, FORMIC ACID | | Authors: | Freedman, S.J, Furie, B.C, Furie, B, Baleja, J.D. | | Deposit date: | 1995-02-26 | | Release date: | 1995-07-10 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the metal-free gamma-carboxyglutamic acid-rich membrane binding region of factor IX by two-dimensional NMR spectroscopy.
J.Biol.Chem., 270, 1995
|
|
1CLD
 
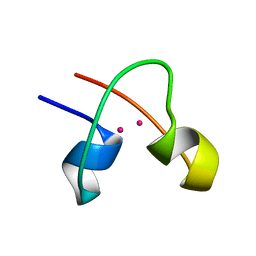 | | DNA-binding protein | | Descriptor: | CADMIUM ION, CD2-LAC9 | | Authors: | Gardner, K.H, Coleman, J.E. | | Deposit date: | 1995-06-06 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Kluyveromyces lactis LAC9 Cd2 Cys6 DNA-binding domain.
Nat.Struct.Biol., 2, 1995
|
|
1CM1
 
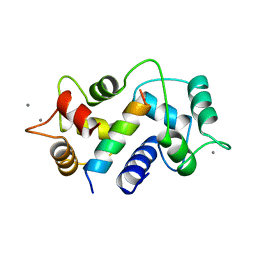 | | MOTIONS OF CALMODULIN-SINGLE-CONFORMER REFINEMENT | | Descriptor: | CALCIUM ION, CALMODULIN, CALMODULIN-DEPENDENT PROTEIN KINASE II-ALPHA | | Authors: | Wall, M.E, Phillips Jr, G.N. | | Deposit date: | 1997-09-23 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Motions of calmodulin characterized using both Bragg and diffuse X-ray scattering.
Structure, 5, 1997
|
|
1COV
 
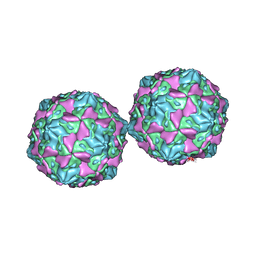 | | COXSACKIEVIRUS B3 COAT PROTEIN | | Descriptor: | COXSACKIEVIRUS COAT PROTEIN, MYRISTIC ACID, PALMITIC ACID | | Authors: | Muckelbauer, J.K, Rossmann, M.G. | | Deposit date: | 1994-10-19 | | Release date: | 1996-03-08 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure determination of coxsackievirus B3 to 3.5 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1CM4
 
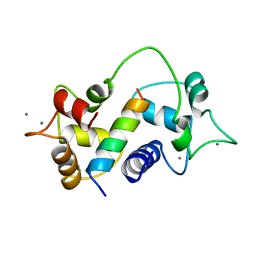 | | Motions of calmodulin-four-conformer refinement | | Descriptor: | CALCIUM ION, CALMODULIN, CALMODULIN-DEPENDENT PROTEIN KINASE II-ALPHA | | Authors: | Wall, M.E, Phillips Jr, G.N. | | Deposit date: | 1997-09-23 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Motions of calmodulin characterized using both Bragg and diffuse X-ray scattering.
Structure, 5, 1997
|
|
1CL1
 
 | |
1CNY
 
 | |
1COL
 
 | | REFINED STRUCTURE OF THE PORE-FORMING DOMAIN OF COLICIN A AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | COLICIN A | | Authors: | Parker, M.W, Postma, J.P.M, Pattus, F, Tucker, A.D, Tsernoglou, D. | | Deposit date: | 1991-07-06 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Refined structure of the pore-forming domain of colicin A at 2.4 A resolution.
J.Mol.Biol., 224, 1992
|
|
1CNS
 
 | |
1CNO
 
 | | STRUCTURE OF PSEUDOMONAS NAUTICA CYTOCHROME C552, BY MAD METHOD | | Descriptor: | CYTOCHROME C552, GLYCEROL, HEME C | | Authors: | Brown, K, Nurizzo, D, Cambillau, C. | | Deposit date: | 1998-08-03 | | Release date: | 1999-07-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MAD structure of Pseudomonas nautica dimeric cytochrome c552 mimicks the c4 Dihemic cytochrome domain association.
J.Mol.Biol., 289, 1999
|
|
1DLH
 
 | |
1DM4
 
 | | SER195ALA MUTANT OF HUMAN THROMBIN COMPLEXED WITH FIBRINOPEPTIDE A (7-16) | | Descriptor: | PROTEIN (ALPHA THROMBIN:LIGHT CHAIN), PROTEIN (FIBRINOPEPTIDE), PROTEIN (MUTANT ALPHA THROMBIN:HEAVY CHAIN) | | Authors: | Krishnan, R, Sadler, E.J, Tulinsky, A. | | Deposit date: | 1999-12-13 | | Release date: | 2000-01-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Ser195Ala mutant of human alpha--thrombin complexed with fibrinopeptide A(7--16): evidence for residual catalytic activity.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1DMW
 
 | | CRYSTAL STRUCTURE OF DOUBLE TRUNCATED HUMAN PHENYLALANINE HYDROXYLASE WITH BOUND 7,8-DIHYDRO-L-BIOPTERIN | | Descriptor: | 7,8-DIHYDROBIOPTERIN, FE (III) ION, PHENYLALANINE HYDROXYLASE | | Authors: | Erlandsen, H, Stevens, R.C, Flatmark, T. | | Deposit date: | 1999-12-15 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and site-specific mutagenesis of pterin-bound human phenylalanine hydroxylase.
Biochemistry, 39, 2000
|
|
1DM6
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH N-(4-CHLOROPHENYL)-N'-HYDROXYGUANIDINE (H4B FREE) | | Descriptor: | ACETATE ION, CACODYLATE ION, N-(CHLOROPHENYL)-N'-HYDROXYGUANIDINE, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Southan, G.J, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-12-13 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of nitric oxide synthase bound to nitro indazole reveals a novel inactivation mechanism.
Biochemistry, 40, 2001
|
|
