3QJF
 
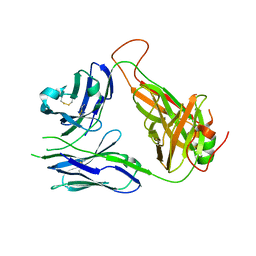 | | Crystal structure of the 2B4 TCR | | Descriptor: | 2B4 alpha chain, 2B4 beta chain | | Authors: | Ely, L.K, Newell, E.W, Davis, M.M, Garcia, K.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k).
J.Immunol., 186, 2011
|
|
4GRP
 
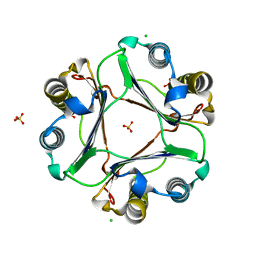 | |
3PRL
 
 | | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125 | | Descriptor: | NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-11-29 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125
To be Published
|
|
3HKW
 
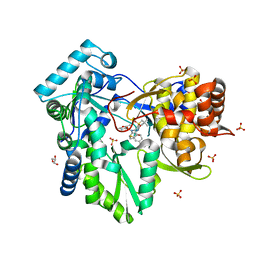 | | HCV NS5B genotype 1a in complex with 1,5 benzodiazepine inhibitor 6 | | Descriptor: | (11S)-10-[(2,5-dimethyl-1,3-oxazol-4-yl)carbonyl]-11-{2-fluoro-4-[(2-methylprop-2-en-1-yl)oxy]phenyl}-3,3-dimethyl-2,3,4,5,10,11-hexahydrothiopyrano[3,2-b][1,5]benzodiazepin-6-ol 1,1-dioxide, GLYCEROL, NS5B RNA-dependent RNA polymerase, ... | | Authors: | Nyanguile, O, De Bondt, H.L. | | Deposit date: | 2009-05-26 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1a/1b subtype profiling of nonnucleoside polymerase inhibitors of hepatitis C virus
J.Virol., 84, 2010
|
|
1XBF
 
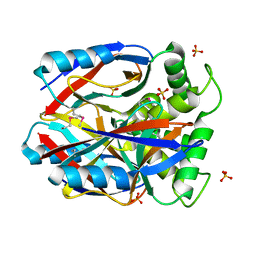 | | X-RAY STRUCTURE NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET CAR10 FROM C. ACETOBUTYLICUM | | Descriptor: | Clostridium acetobutylicum Q97KL0, SULFATE ION | | Authors: | Kuzin, A.P, Chen, Y, Vorobiev, S, Yong, W, Acton, T, Ho, C.-K, Conover, K, Cooper, B, Ciano, M, Xiao, R, Montelione, G, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-08-30 | | Release date: | 2004-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-RAY STRUCTURE NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET CAR10 FROM C. ACETOBUTYLICUM
To be published
|
|
3IQU
 
 | |
4FK6
 
 | | JAK1 kinase (JH1 domain) in complex with compound 72 | | Descriptor: | N-({1-[(1R,2R,4S)-bicyclo[2.2.1]hept-2-yl]-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridin-2-yl}methyl)methanesulfonamide, Tyrosine-protein kinase JAK1 | | Authors: | Eigenbrot, C, Steffek, M. | | Deposit date: | 2012-06-12 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based discovery of C-2 substituted imidazo-pyrrolopyridine JAK1 inhibitors with improved selectivity over JAK2.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3ORV
 
 | |
4M6Y
 
 | | Mutant structure of methyltransferase from Streptomyces hygroscopicus complexed with S-adenosyl-L-homocysteine and methylphenylpyruvic acid | | Descriptor: | (2R)-2-hydroxy-3-phenylpropanoic acid, (3R)-2-oxo-3-phenylbutanoic acid, CALCIUM ION, ... | | Authors: | Liu, Y.C, Zou, X.W, Chan, H.C, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-12 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of a nonhaem-iron SAM-dependent C-methyltransferase and its engineering to a hydratase and an O-methyltransferase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4M73
 
 | | Mutant structure of methyltransferase from Streptomyces hygroscopicus | | Descriptor: | (2R)-2-hydroxy-3-phenylpropanoic acid, (2R,3R)-2-hydroxy-3-methoxy-3-phenylpropanoic acid, CALCIUM ION, ... | | Authors: | Liu, Y.C, Zou, X.W, Chan, H.C, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-12 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of a nonhaem-iron SAM-dependent C-methyltransferase and its engineering to a hydratase and an O-methyltransferase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3IQJ
 
 | | Crystal Structure of human 14-3-3 sigma in Complex with Raf1 peptide (10mer) | | Descriptor: | 10-mer peptide from RAF proto-oncogene serine/threonine-protein kinase, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Ottmann, C, Weyand, M. | | Deposit date: | 2009-08-20 | | Release date: | 2010-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Impaired binding of 14-3-3 to C-RAF in Noonan syndrome suggests new approaches in diseases with increased Ras signaling.
Mol.Cell.Biol., 30, 2010
|
|
2XLD
 
 | |
5CCP
 
 | | HISTIDINE 52 IS A CRITICAL RESIDUE FOR RAPID FORMATION OF CYTOCHROME C PEROXIDASE COMPOUND I | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Brown, K, Shaw, A, Miller, M.A, Kraut, J. | | Deposit date: | 1993-06-07 | | Release date: | 1993-10-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Histidine 52 is a critical residue for rapid formation of cytochrome c peroxidase compound I.
Biochemistry, 32, 1993
|
|
1OG7
 
 | | Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide sakacin P. | | Descriptor: | BACTERIOCIN SAKACIN P | | Authors: | Uteng, M, Hauge, H.H, Markwick, P.R, Fimland, G, Mantzilas, D, Nissen-Meyer, J, Muhle-Goll, C. | | Deposit date: | 2003-04-25 | | Release date: | 2003-09-22 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure in Lipid Micelles of the Pediocin-Like Antimicrobial Peptide Sakacin P and a Sakacin P Variant that is Structurally Stabilized by an Inserted C-Terminal Disulfide Bridge
Biochemistry, 42, 2003
|
|
1OHN
 
 | | Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide sakacin P. | | Descriptor: | BACTERIOCIN SAKACIN P | | Authors: | Uteng, M, Hauge, H.H, Markwick, P.R, Fimland, G, Mantzilas, D, Nissen-Meyer, J, Muhle-Goll, C. | | Deposit date: | 2003-05-28 | | Release date: | 2003-09-22 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure in Lipid Micelles of the Pediocin-Like Antimicrobial Peptide Sakacin P and a Sakacin P Variant that is Structurally Stabilized by an Inserted C-Terminal Disulfide Bridge
Biochemistry, 42, 2003
|
|
1NS3
 
 | | STRUCTURE OF HCV PROTEASE (BK STRAIN) | | Descriptor: | NS3 PROTEASE, NS4A PEPTIDE, ZINC ION | | Authors: | Yan, Y, Munshi, S, Chen, Z. | | Deposit date: | 1997-04-05 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form.
Protein Sci., 7, 1998
|
|
1OAE
 
 | | Crystal structure of the reduced form of cytochrome c" from Methylophilus methylotrophus | | Descriptor: | CYTOCHROME C", GLYCEROL, HEME C, ... | | Authors: | Enguita, F.J, Grenha, R, Santos, H, Carrondo, M.A. | | Deposit date: | 2003-01-09 | | Release date: | 2004-03-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Evidence for a Proton Transfer Pathway Coupled with Haem Reduction of Cytochrome C" from Methylophilus Methylotrophus.
J.Biol.Inorg.Chem., 11, 2006
|
|
3BRP
 
 | | Crystal Structure of C-Phycocyanin from Galdieria sulphuraria | | Descriptor: | BILIVERDINE IX ALPHA, C-phycocyanin alpha chain, C-phycocyanin beta chain | | Authors: | Fromme, R. | | Deposit date: | 2007-12-21 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of C-Phycocyanin from Galdieria sulphuraria at 1.85 A
To be Published
|
|
4DEG
 
 | | Crystal structure of c-Met in complex with triazolopyridazine inhibitor 2 | | Descriptor: | 7-methoxy-N-{[6-(3-methyl-1,2-thiazol-5-yl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]methyl}-1,5-naphthyridin-4-amine, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Bellon, S.F, Long, A.M. | | Deposit date: | 2012-01-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and optimization of a potent and selective triazolopyridinone series of c-Met inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4M71
 
 | | Mutant structure of methyltransferase from Streptomyces hygroscopicus complexed with S-adenosyl-L-homocysteine and methylphenylpyruvic acid | | Descriptor: | (2R)-2-hydroxy-3-phenylpropanoic acid, (3R)-2-oxo-3-phenylbutanoic acid, CALCIUM ION, ... | | Authors: | Liu, Y.C, Zou, X.W, Chan, H.C, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-12 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure and mechanism of a nonhaem-iron SAM-dependent C-methyltransferase and its engineering to a hydratase and an O-methyltransferase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5B3I
 
 | | Homo-dimeric structure of cytochrome c' from Thermophilic Hydrogenophilus thermoluteolus | | Descriptor: | Cytochrome c prime, HEME C | | Authors: | Fujii, S, Oki, H, Kawahara, K, Yamane, D, Yamanaka, M, Maruno, T, Kobayashi, Y, Masanari, M, Wakai, S, Nishihara, H, Ohkubo, T, Sambongi, Y. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and functional insights into thermally stable cytochrome c' from a thermophile
Protein Sci., 26, 2017
|
|
7X8K
 
 | | Arabidopsis GDP-D-mannose pyrophosphorylase (VTC1) structure (product-bound) | | Descriptor: | CITRATE ANION, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, Mannose-1-phosphate guanylyltransferase 1, ... | | Authors: | Zhao, S, Zhang, C, Liu, L. | | Deposit date: | 2022-03-13 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Arabidopsis thaliana GDP-D-Mannose Pyrophosphorylase VITAMIN C DEFECTIVE 1.
Front Plant Sci, 13, 2022
|
|
3RHH
 
 | | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125 complexed with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125 complexed with NADP
To be Published
|
|
5C9M
 
 | | The structure of oxidized rat cytochrome c (T28A) at 1.362 angstroms resolution. | | Descriptor: | Cytochrome c, somatic, HEME C, ... | | Authors: | Edwards, B.F.P, Mahapatra, G, Vaishnav, A.A, Brunzelle, J.S, Huttemann, M. | | Deposit date: | 2015-06-28 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.362 Å) | | Cite: | The structure of oxidized rat cytochrome c (T28A) at 1.362 angstroms resolution.
To Be Published
|
|
1Q31
 
 | | Crystal Structure of the Tobacco Etch Virus Protease C151A mutant | | Descriptor: | BETA-MERCAPTOETHANOL, Nuclear inclusion protein A | | Authors: | Nunn, C.M, Djordjevic, S, George, R.R, Urquhart, G.T, Chao, L.H, Tsuchiya, Y. | | Deposit date: | 2003-07-28 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of tobacco etch virus protease shows the protein C terminus bound within the active site.
J.Mol.Biol., 350, 2005
|
|
