2VRO
 
 | | Crystal structure of aldehyde dehydrogenase from Burkholderia xenovorans LB400 | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ALDEHYDE DEHYDROGENASE, HEXAETHYLENE GLYCOL, ... | | Authors: | Bains, J, Boulanger, M.J. | | Deposit date: | 2008-04-09 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Characterization of a Novel Aldehyde Dehydrogenase Encoded by the Benzoate Oxidation (Box) Pathway in Burkholderia Xenovorans Lb400
J.Mol.Biol., 379, 2008
|
|
3CB7
 
 | | The crystallographic structure of the digestive lysozyme 2 from Musca domestica at 1.9 Ang. | | Descriptor: | ACETIC ACID, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Cancado, F.C, Valerio, A.A, Marana, S.R, Barbosa, J.A.R.G. | | Deposit date: | 2008-02-21 | | Release date: | 2009-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystallographic structure of the digestive lysozyme 2 from Musca domestica at 1.9 Ang.
To be Published
|
|
3AAX
 
 | | Crystal structure of probable thiosulfate sulfurtransferase cysa3 (RV3117) from Mycobacterium tuberculosis: monoclinic FORM | | Descriptor: | Putative thiosulfate sulfurtransferase | | Authors: | Sankaranarayanan, R, Witholt, S.J, Cherney, M.M, Garen, C.R, Cherney, L.T, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of probable thiosulfate sulfurtransferase CysA3 (Rv3117) from Mycobacterium tuberculosis
To be Published
|
|
3IMV
 
 | |
3AAY
 
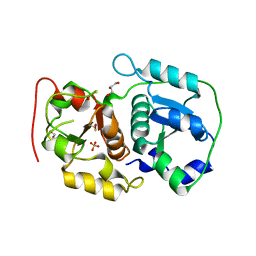 | | Crystal structure of probable thiosulfate sulfurtransferase CYSA3 (RV3117) from Mycobacterium tuberculosis: orthorhombic form | | Descriptor: | GLYCEROL, Putative thiosulfate sulfurtransferase, SULFATE ION | | Authors: | Sankaranarayanan, R, Witholt, S.J, Cherney, M.M, Garen, C.R, Cherney, L.T, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of probable thiosulfate sulfurtransferase CysA3 (Rv3117) from Mycobacterium tuberculosis
To be Published
|
|
2QI0
 
 | |
2QI7
 
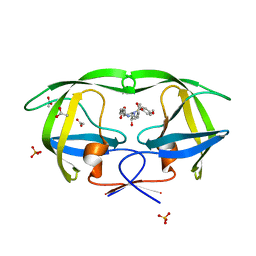 | | Crystal structure of protease inhibitor, MIT-2-AD86 in complex with wild type HIV-1 protease | | Descriptor: | ACETATE ION, N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{[(4-METHOXYPHENYL)SULFONYL][(2S)-2-METHYLBUTYL]AMINO}PROPYL]-4-OXOHEXANAMIDE, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
3D7P
 
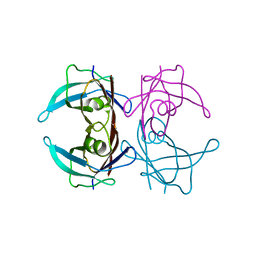 | | Crystal structure of human Transthyretin (TTR) at pH 4.0 | | Descriptor: | Transthyretin | | Authors: | Palaninathan, S.K, Mohamedmohaideen, N.N, Snee, W.C, Kelly, J.W, Sacchettini, J.C. | | Deposit date: | 2008-05-21 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural insight into pH-induced conformational changes within the native human transthyretin tetramer.
J.Mol.Biol., 382, 2008
|
|
3CBR
 
 | | Crystal structure of human Transthyretin (TTR) at pH3.5 | | Descriptor: | Transthyretin | | Authors: | Mohamedmohaideen, N.N, Palaninathan, S.K, Snee, W.C, Kelly, J.W, C Sacchettini, J. | | Deposit date: | 2008-02-22 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into pH-induced conformational changes within the native human transthyretin tetramer.
J.Mol.Biol., 382, 2008
|
|
2RL3
 
 | | Crystal structure of the OXA-10 W154H mutant at pH 7 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase PSE-2, GLYCEROL, ... | | Authors: | Vercheval, L, Kerff, F, Herman, R, Sauvage, E, Guiet, R, Charlier, P, Frere, J.-M, Galleni, M. | | Deposit date: | 2007-10-18 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
2PB0
 
 | | Structure of biosynthetic N-acetylornithine aminotransferase from Salmonella typhimurium: studies on substrate specificity and inhibitor binding | | Descriptor: | 1,2-ETHANEDIOL, Acetylornithine/succinyldiaminopimelate aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Rajaram, V, Ratna Prasuna, P, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2007-03-28 | | Release date: | 2007-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of biosynthetic N-acetylornithine aminotransferase from Salmonella typhimurium: Studies on substrate specificity and inhibitor binding
Proteins, 70, 2007
|
|
5U28
 
 | | BRD4 first bromodomain (BD1) in complex with dual PI3 kinase inhibitor SF2523 | | Descriptor: | 3-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-(morpholin-4-yl)-7H-thieno[3,2-b]pyran-7-one, Bromodomain-containing protein 4, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Andrews, F.H, Kutateladze, T.G. | | Deposit date: | 2016-11-30 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Dual-activity PI3K-BRD4 inhibitor for the orthogonal inhibition of MYC to block tumor growth and metastasis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5U2F
 
 | |
5U2E
 
 | |
3USR
 
 | | Structure of Y194F glycogenin mutant truncated at residue 270 | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycogenin-1 | | Authors: | Issoglio, F.M, Carrizo, M.E, Romero, J.M, Curtino, J.A. | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanisms of monomeric and dimeric glycogenin autoglucosylation.
J.Biol.Chem., 287, 2012
|
|
3USQ
 
 | | Structure of D159S/Y194F glycogenin mutant truncated at residue 270 | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycogenin-1 | | Authors: | Issoglio, F.M, Carrizo, M.E, Romero, J.M, Curtino, J.A. | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanisms of monomeric and dimeric glycogenin autoglucosylation.
J.Biol.Chem., 287, 2012
|
|
4B1C
 
 | | New Aminoimidazoles as BACE-1 Inhibitors: From Rational Design to Ab- lowering in Brain | | Descriptor: | (2R)-2-cyclopropyl-5-methyl-2-[3-(5-prop-1-yn-1-ylpyridin-3-yl)phenyl]-2H-imidazol-4-amine, BETA-SECRETASE 1, DIMETHYL SULFOXIDE | | Authors: | Rahm, F, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, von Berg, S, von Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Gravenfors, Y. | | Deposit date: | 2012-07-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New aminoimidazoles as beta-secretase (BACE-1) inhibitors showing amyloid-beta (A beta ) lowering in brain.
J. Med. Chem., 55, 2012
|
|
4AO6
 
 | | Native structure of a novel cold-adapted esterase from an Arctic intertidal metagenomic library | | Descriptor: | ESTERASE | | Authors: | Fu, J, Leiros, H.-K.S, Pascale, D.d, Johnson, K.A, Blencke, H.M, Landfald, B. | | Deposit date: | 2012-03-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional and Structural Studies of a Novel Cold-Adapted Esterase from an Arctic Intertidal Metagenomic Library.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
4AO7
 
 | | Zinc bound structure of a novel cold-adapted esterase from an Arctic intertidal metagenomic library | | Descriptor: | ESTERASE, ZINC ION | | Authors: | Fu, J, Leiros, H.-K.S, Pascale, D.d, Johnson, K.A, Blencke, H.M, Landfald, B. | | Deposit date: | 2012-03-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Functional and Structural Studies of a Novel Cold-Adapted Esterase from an Arctic Intertidal Metagenomic Library.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
4C1K
 
 | | Crystal structure of pyrococcus furiosus 3-deoxy-D-arabino- heptulosonate 7-phosphate synthase | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOHEPTONATE ALDOLASE, CADMIUM ION, CARBONATE ION, ... | | Authors: | Nazmi, A.R, Schofield, L.R, Dobson, R.C.J, Jameson, G.B, Parker, E.J. | | Deposit date: | 2013-08-13 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Destabilization of the Homotetrameric Assembly of 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase from the Hyperthermophile Pyrococcus Furiosus Enhances Enzymatic Activity
J.Mol.Biol., 426, 2014
|
|
4AO8
 
 | | PEG-bound complex of a novel cold-adapted esterase from an Arctic intertidal metagenomic library | | Descriptor: | DI(HYDROXYETHYL)ETHER, ESTERASE | | Authors: | Fu, J, Leiros, H.-K.S, Pascale, D.d, Johnson, K.A, Blencke, H.M, Landfald, B. | | Deposit date: | 2012-03-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Functional and Structural Studies of a Novel Cold-Adapted Esterase from an Arctic Intertidal Metagenomic Library.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
4C1L
 
 | | Crystal structure of pyrococcus furiosus 3-deoxy-D-arabino- heptulosonate 7-phosphate synthase I181D interface mutant | | Descriptor: | 2-dehydro-3-deoxyphosphoheptonate aldolase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Nazmi, A.R, Schofield, L.R, Dobson, R.C.J, Jameson, G.B, Parker, E.J. | | Deposit date: | 2013-08-13 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Destabilization of the homotetrameric assembly of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase from the hyperthermophile Pyrococcus furiosus enhances enzymatic activity.
J. Mol. Biol., 426, 2014
|
|
3U1H
 
 | |
3V8Z
 
 | | Structure of apo-glycogenin truncated at residue 270 complexed with UDP | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycogenin-1, ... | | Authors: | Carrizo, M.E, Romero, J.M, Issoglio, F.M, Curtino, J.A. | | Deposit date: | 2011-12-23 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical insight into glycogenin inactivation by the glycogenosis-causing T82M mutation.
Febs Lett., 586, 2012
|
|
3V90
 
 | | Structure of T82M glycogenin mutant truncated at residue 270 | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycogenin-1 | | Authors: | Carrizo, M.E, Romero, J.M, Issoglio, F.M, Curtino, J.A. | | Deposit date: | 2011-12-23 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical insight into glycogenin inactivation by the glycogenosis-causing T82M mutation.
Febs Lett., 586, 2012
|
|
