6R44
 
 | |
8HOC
 
 | | Cryo-EM structure of ligand histamine-bound Histamine H4 receptor Gi complex | | Descriptor: | 2-(1~{H}-imidazol-5-yl)ethyl carbamimidothioate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tang, W.Q, Sun, X.Y, Li, F.H, Wang, J.Y. | | Deposit date: | 2022-12-09 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure insights into Histamine H4 receptor activation by an endogenous ligand histamine and agonist imetit
To Be Published
|
|
4TYI
 
 | | Structural analysis of the human Fibroblast Growth Factor Receptor 4 | | Descriptor: | 4-amino-5-fluoro-3-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]quinolin-2(1H)-one, Fibroblast growth factor receptor 4 | | Authors: | Lesca, E, Lammens, A, Huber, R, Augustin, M. | | Deposit date: | 2014-07-08 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural analysis of the human fibroblast growth factor receptor 4 kinase.
J.Mol.Biol., 426, 2014
|
|
8HN8
 
 | | Cryo-EM structure of ligand histamine-bound Histamine H4 receptor Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Tang, W.Q, Sun, X.Y, Li, F.H, Wang, J.Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure insights into Histamine H4 receptor activation by an endogenous ligand histamine and agonist imetit
To Be Published
|
|
6R4R
 
 | | Cryo-EM Structure of the PI3-Kinase SH3 Domain Amyloid Fibril | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha | | Authors: | Roeder, C, Vettore, N, Mangels, L.N, Gremer, L, Ravelli, R.B.G, Willbold, D, Hoyer, W, Buell, A.K, Schroder, G.F. | | Deposit date: | 2019-03-23 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomic structure of PI3-kinase SH3 amyloid fibrils by cryo-electron microscopy.
Nat Commun, 10, 2019
|
|
6R9O
 
 | | Structure of Saccharomyces cerevisiae apo Pan2 pseudoubiquitin hydrolase-RNA exonuclease (UCH-Exo) module in complex with AAGGA RNA | | Descriptor: | AAGGA RNA, PAN2-PAN3 deadenylation complex catalytic subunit PAN2 | | Authors: | Tang, T.T.L, Stowell, J.A.W, Hill, C.H, Passmore, L.A. | | Deposit date: | 2019-04-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.319 Å) | | Cite: | The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6R9T
 
 | | Cryo-EM structure of autoinhibited human talin-1 | | Descriptor: | Talin-1 | | Authors: | Dedden, D, Schumacher, S, Zacharias, M, Biertumpfel, C, Mizuno, N. | | Deposit date: | 2019-04-04 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | The Architecture of Talin1 Reveals an Autoinhibition Mechanism.
Cell, 179, 2019
|
|
6R6B
 
 | | Structure of the core Shigella flexneri type III secretion system export gate complex SctRST (Spa24/Spa9/Spa29). | | Descriptor: | Surface presentation of antigens protein SpaP, Surface presentation of antigens protein SpaQ, Surface presentation of antigens protein SpaR | | Authors: | Johnson, S, Kuhlen, L, Deme, J.C, Abrusci, P, Lea, S.M. | | Deposit date: | 2019-03-26 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The Structure of an Injectisome Export Gate Demonstrates Conservation of Architecture in the Core Export Gate between Flagellar and Virulence Type III Secretion Systems.
Mbio, 10, 2019
|
|
6RC8
 
 | |
6RD2
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-1]-TEDEL-NH2 | | Descriptor: | (3~{S},7~{R},10~{R},13~{S})-4-[(3~{S},6~{R},8~{a}~{S})-1'-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8~{a}-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonyl-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxylic acid, GLYCEROL, NITRATE ION, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2019-04-12 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6RD9
 
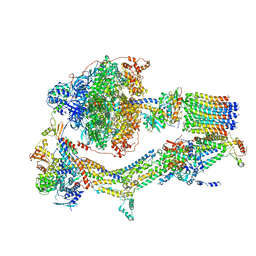 | | CryoEM structure of Polytomella F-ATP synthase, Primary rotary state 1, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
7VBG
 
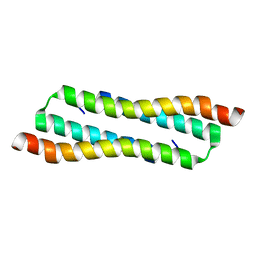 | | N Terminal Domain of PRC1 | | Descriptor: | Protein regulator of cytokinesis 1 | | Authors: | Tan, F, Xia, B. | | Deposit date: | 2021-08-31 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | N Terminal Domain of PRC1
To Be Published
|
|
6RDT
 
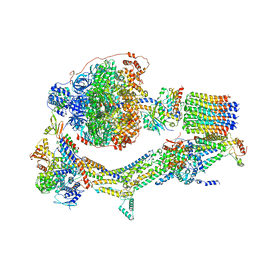 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1E, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6REA
 
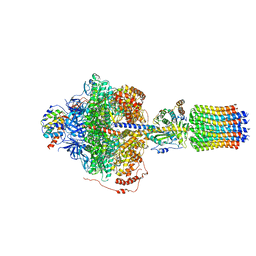 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 2D, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RES
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 3C, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
4UA6
 
 | |
4UAM
 
 | | 1.8 Angstrom crystal structure of IMP-1 metallo-beta-lactamase with a mixed iron-zinc center in the active site | | Descriptor: | CITRATE ANION, FE (III) ION, IMP-1 metallo-beta-lactamase, ... | | Authors: | Carruthers, T.J, Carr, P.D, Jackson, C.J, Otting, G. | | Deposit date: | 2014-08-11 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Iron(III) Located in the Dinuclear Metallo-beta-Lactamase IMP-1 by Pseudocontact Shifts.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6RAV
 
 | | Complement factor B protease domain in complex with the reversible inhibitor 4-((2S,4S)-4-ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic acid | | Descriptor: | 4-[(2~{S},4~{S})-4-ethoxy-1-[(5-methoxy-7-methyl-1~{H}-indol-4-yl)methyl]piperidin-2-yl]benzoic acid, Complement factor B, SULFATE ION, ... | | Authors: | Adams, C.M, Sellner, H, Ehara, T, Mac Sweeney, A, Crowley, M, Anderson, K, Karki, R, Mainolfi, N, Valeur, E, Sirockin, F, Gerhartz, B, Erbel, P, Hughes, N, Smith, T.M, Cumin, F, Argikar, U, Mogi, M, Sedrani, R, Wiesmann, C, Jaffee, B, Maibaum, J, Flohr, S, Harrison, R, Eder, J. | | Deposit date: | 2019-04-08 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small-molecule factor B inhibitor for the treatment of complement-mediated diseases.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4TZY
 
 | |
6RDB
 
 | | CryoEM structure of Polytomella F-ATP synthase, Primary rotary state 1, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RDP
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1C, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RDZ
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 2A, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6REF
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 3B, monomer-masked refinement | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6R7Y
 
 | | CryoEM structure of calcium-bound human TMEM16K / Anoctamin 10 in detergent (low Ca2+, closed form) | | Descriptor: | Anoctamin-10, CALCIUM ION | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
6RBD
 
 | | State 1 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Mitterer, V, Ferreira-Cerca, S, Murat, G, Enne, T, Rinaldi, D, Weigl, S, Omanic, H, Gleizes, P.E, Kressler, D, Pertschy, B, Plisson-Chastang, C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism.
Nat Commun, 10, 2019
|
|
