3K8L
 
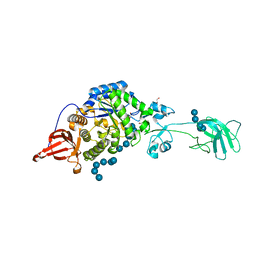 | |
3RDR
 
 | | Structure of the catalytic domain of XlyA | | Descriptor: | CHLORIDE ION, N-acetylmuramoyl-L-alanine amidase XlyA, ZINC ION | | Authors: | Low, L.Y, Liddington, R.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of net charge on catalytic domain and influence of cell wall binding domain on bactericidal activity, specificity, and host range of phage lysins.
J.Biol.Chem., 286, 2011
|
|
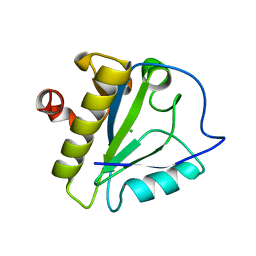
3RTY
 
 | | Structure of an Enclosed Dimer Formed by The Drosophila Period Protein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Period circadian protein | | Authors: | King, H.A, Hoelz, A, Crane, B.R, Young, M.W. | | Deposit date: | 2011-05-04 | | Release date: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of an enclosed dimer formed by the Drosophila period protein.
J.Mol.Biol., 413, 2011
|
|
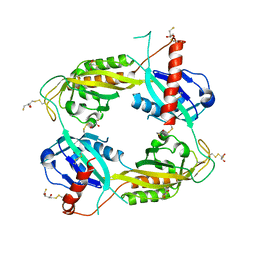
1SJF
 
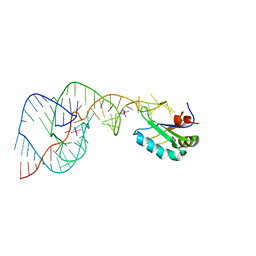 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaion, in Cobalt Hexammine solution | | Descriptor: | COBALT HEXAMMINE(III), Hepatitis Delta virus ribozyme, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A conformational switch controls hepatitis delta virus ribozyme catalysis.
Nature, 429, 2004
|
|
2QUW
 
 | |
7M50
 
 | |
2QG6
 
 | |
2M4Q
 
 | | NMR structure of E. coli ribosomela decoding site with apramycin | | Descriptor: | APRAMYCIN, RNA (27-MER) | | Authors: | Puglisi, J.D, Tsai, A, Marshall, R, Viani, E. | | Deposit date: | 2013-02-10 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The impact of aminoglycosides on the dynamics of translation elongation.
Cell Rep, 3, 2013
|
|
4GLA
 
 | | OBody NL8 bound to hen egg-white lysozyme | | Descriptor: | Lysozyme C, OBody NL8 | | Authors: | Steemson, J.D. | | Deposit date: | 2012-08-14 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Tracking Molecular Recognition at the Atomic Level with a New Protein Scaffold Based on the OB-Fold.
Plos One, 9, 2014
|
|
6YBT
 
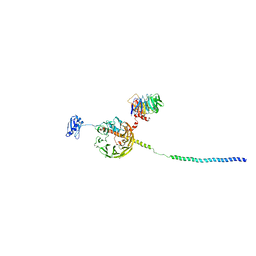 | | Structure of a human 48S translational initiation complex - eIF3bgi | | Descriptor: | Eukaryotic translation initiation factor 3 subunit A, Eukaryotic translation initiation factor 3 subunit B, Eukaryotic translation initiation factor 3 subunit I | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-17 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
6YNN
 
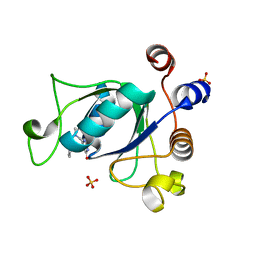 | | Crystal structure of YTHDC1 with compound DHU_DC1_135 | | Descriptor: | 6-[[(2-chloranyl-6-fluoranyl-phenyl)methyl-methyl-amino]methyl]-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
1JFI
 
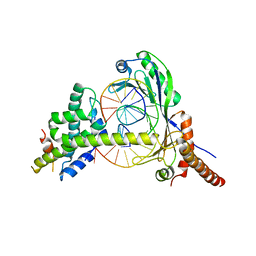 | | Crystal Structure of the NC2-TBP-DNA Ternary Complex | | Descriptor: | 5'-D(*G*GP*AP*GP*CP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*CP*CP*AP*A)-3', 5'-D(*TP*TP*GP*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*TP*CP*C)-3', TATA-BOX-BINDING PROTEIN (TBP), ... | | Authors: | Kamada, K, Shu, F, Chen, H, Malik, S, Stelzer, G, Roeder, R.G, Meisterernst, M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-06-20 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex.
Cell(Cambridge,Mass.), 106, 2001
|
|
1NJN
 
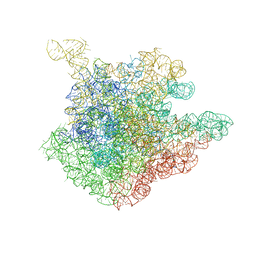 | | The crystal structure of the 50S Large ribosomal subunit from Deinococcus radiodurans complexed with the antibiotic sparsomycin | | Descriptor: | 23S ribosomal RNA, SPARSOMYCIN | | Authors: | Bashan, A, Agmon, I, Zarivatch, R, Schluenzen, F, Harms, J.M, Berisio, R, Bartels, H, Hansen, H.A, Yonath, A. | | Deposit date: | 2003-01-02 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis of the ribosomal machinery for Peptide bond formation,
translocation, and nascent chain progression
Mol.Cell, 11, 2003
|
|
6YNO
 
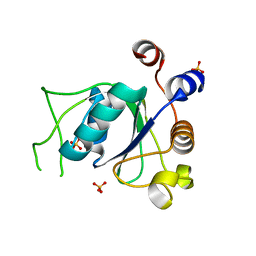 | | Crystal structure of YTHDC1 with compound DHU_DC1_139 | | Descriptor: | 6-[[methyl-[(1-phenylpyrazol-3-yl)methyl]amino]methyl]-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
481D
 
 | |
2W9J
 
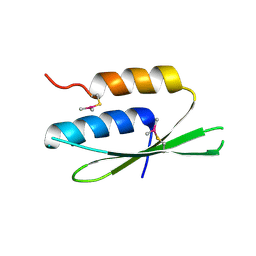 | | The crystal structure of SRP14 from the Schizosaccharomyces pombe signal recognition particle | | Descriptor: | SIGNAL RECOGNITION PARTICLE SUBUNIT SRP14 | | Authors: | Brooks, M.A, Ravelli, R.B.G, McCarthy, A.A, Strub, K, Cusack, S. | | Deposit date: | 2009-01-25 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Srp14 from the Schizosaccharomyces Pombe Signal Recognition Particle.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6Z45
 
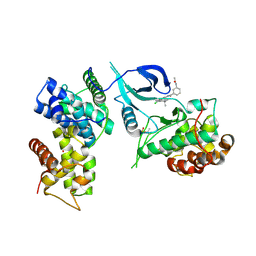 | | CDK9-Cyclin-T1 complex bound by compound 24 | | Descriptor: | (1~{S},3~{R})-3-acetamido-~{N}-[5-chloranyl-4-(5,5-dimethyl-4,6-dihydropyrrolo[1,2-b]pyrazol-3-yl)pyridin-2-yl]cyclohexane-1-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cyclin-T1, ... | | Authors: | Ferguson, A, Collie, G.W. | | Deposit date: | 2020-05-22 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Discovery of AZD4573, a Potent and Selective Inhibitor of CDK9 That Enables Short Duration of Target Engagement for the Treatment of Hematological Malignancies.
J.Med.Chem., 63, 2020
|
|
5D6Y
 
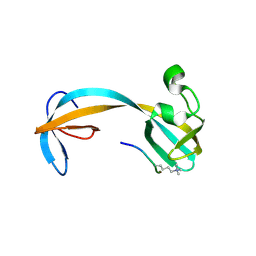 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A complexed with histone H3K23me3 | | Descriptor: | Lysine-specific demethylase 4A, peptide H3K23me3 (19-28) | | Authors: | Wang, F, Su, Z, Miller, M.D, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-08-13 | | Release date: | 2016-02-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
2FYR
 
 | |
6ZD9
 
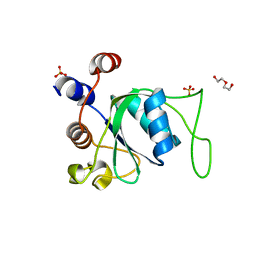 | | Crystal structure of YTHDC1 apo purified using GST tag | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Caflisch, A. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6ZD7
 
 | |
5ES3
 
 | | Co-crystal structure of LDH liganded with oxamate | | Descriptor: | L-lactate dehydrogenase A chain, OXAMIC ACID | | Authors: | Nowicki, M.W, Wear, M.A, McNae, I.W, Blackburn, E.A. | | Deposit date: | 2015-11-16 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Streamlined, Automated Protocol for the Production of Milligram Quantities of Untagged Recombinant Rat Lactate Dehydrogenase A Using AKTAxpressTM.
Plos One, 10, 2015
|
|
6ZD4
 
 | | Crystal structure of YTHDC1 S378A mutant | | Descriptor: | SULFATE ION, YTH domain containing 1 | | Authors: | Bedi, R.K, Li, Y, Caflisch, A. | | Deposit date: | 2020-06-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
4CS1
 
 | |
6ZCN
 
 | | Crystal structure of YTHDC1 with m6A | | Descriptor: | N6-METHYLADENOSINE-5'-MONOPHOSPHATE, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
