1HRD
 
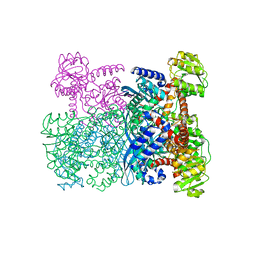 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE | | Authors: | Britton, K.L, Baker, P.J, Stillman, T.J, Rice, D.W. | | Deposit date: | 1996-04-03 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The structure of Pyrococcus furiosus glutamate dehydrogenase reveals a key role for ion-pair networks in maintaining enzyme stability at extreme temperatures.
Structure, 3, 1995
|
|
2KQT
 
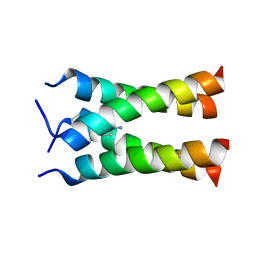 | | Solid-state NMR structure of the M2 transmembrane peptide of the influenza A virus in DMPC lipid bilayers bound to deuterated amantadine | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, M2 protein | | Authors: | Cady, S.D, Schmidt-Rohr, K, Wang, J, Soto, C.S, DeGrado, W.F, Hong, M. | | Deposit date: | 2009-11-18 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-08 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers
Nature, 463, 2010
|
|
1BFC
 
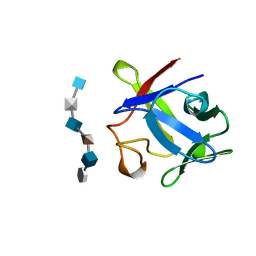 | | BASIC FIBROBLAST GROWTH FACTOR COMPLEXED WITH HEPARIN HEXAMER FRAGMENT | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, BASIC FIBROBLAST GROWTH FACTOR | | Authors: | Faham, S, Rees, D.C. | | Deposit date: | 1995-12-12 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Heparin structure and interactions with basic fibroblast growth factor.
Science, 271, 1996
|
|
2Y32
 
 | | Crystal structure of bradavidin | | Descriptor: | BLR5658 PROTEIN | | Authors: | Leppiniemi, J, Gronroos, T, Johnson, M.S, Kulomaa, M.S, Hytonen, V.P, Airenne, T.T. | | Deposit date: | 2010-12-17 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of Bradavidin - C-Terminal Residues Act as Intrinsic Ligands.
Plos One, 7, 2012
|
|
1APV
 
 | |
2F6I
 
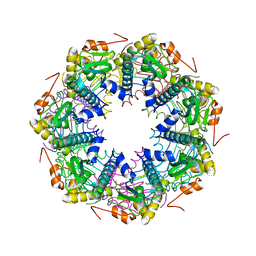 | | Crystal structure of the ClpP protease catalytic domain from Plasmodium falciparum | | Descriptor: | ATP-dependent CLP protease, putative | | Authors: | Mulichak, A, Loppnau, P, Bray, J, Amani, M, Vedadi, M, Wasney, G, Finerty, P, Sundstrom, M, Weigelt, J, Edwards, A, Arrowsmith, C, Hui, R, Plotnikova, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-29 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Clp chaperones and proteases of the human malaria parasite Plasmodium falciparum.
J.Mol.Biol., 404, 2010
|
|
1J88
 
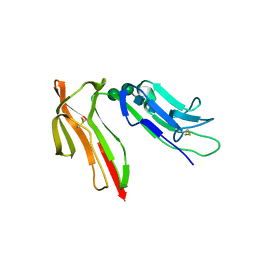 | | HUMAN HIGH AFFINITY FC RECEPTOR FC(EPSILON)RI(ALPHA), TETRAGONAL CRYSTAL FORM 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIGH AFFINITY IMMUNOGLOBULIN EPSILON RECEPTOR ALPHA-SUBUNIT, ... | | Authors: | Garman, S.C, Sechi, S, Kinet, J.P, Jardetzky, T.S. | | Deposit date: | 2001-05-20 | | Release date: | 2001-08-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The analysis of the human high affinity IgE receptor Fc epsilon Ri alpha from multiple crystal forms.
J.Mol.Biol., 311, 2001
|
|
1DD7
 
 | |
1LNC
 
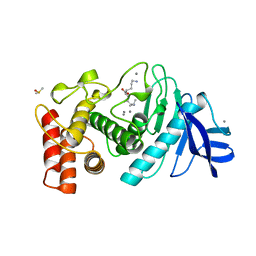 | | A STRUCTURAL ANALYSIS OF METAL SUBSTITUTIONS IN THERMOLYSIN | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, LYSINE, ... | | Authors: | Holland, D.R, Hausrath, A.C, Juers, D, Matthews, B.W. | | Deposit date: | 1994-05-13 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of zinc substitutions in the active site of thermolysin.
Protein Sci., 4, 1995
|
|
1LNA
 
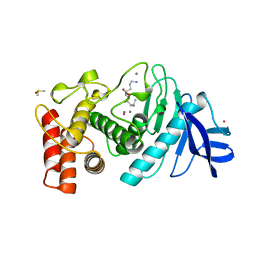 | | A STRUCTURAL ANALYSIS OF METAL SUBSTITUTIONS IN THERMOLYSIN | | Descriptor: | CALCIUM ION, COBALT (II) ION, DIMETHYL SULFOXIDE, ... | | Authors: | Holland, D.R, Hausrath, A.C, Juers, D, Matthews, B.W. | | Deposit date: | 1994-05-13 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of zinc substitutions in the active site of thermolysin.
Protein Sci., 4, 1995
|
|
2Y8N
 
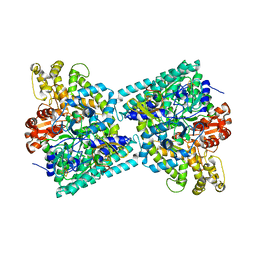 | | Crystal structure of glycyl radical enzyme | | Descriptor: | 4-HYDROXYPHENYLACETATE DECARBOXYLASE LARGE SUBUNIT, 4-HYDROXYPHENYLACETATE DECARBOXYLASE SMALL SUBUNIT, IRON/SULFUR CLUSTER | | Authors: | Martins, B.M, Blaser, M, Feliks, M, Ullmann, G.M, Selmer, T. | | Deposit date: | 2011-02-08 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for a Kolbe-Type Decarboxylation Catalyzed by a Glycyl Radical Enzyme.
J.Am.Chem.Soc., 133, 2011
|
|
2DT1
 
 | | Crystal Structure Of The Complex Of Goat Signalling Protein With Tetrasaccharide At 2.09 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kumar, J, Ethayathulla, A.S, Srivastava, D.B, Singh, N, Sharma, S, Bhushan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2006-07-09 | | Release date: | 2006-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Carbohydrate-binding properties of goat secretory glycoprotein (SPG-40) and its functional implications: structures of the native glycoprotein and its four complexes with chitin-like oligosaccharides
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
1UOR
 
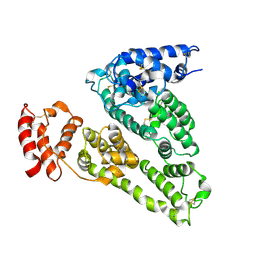 | |
1J83
 
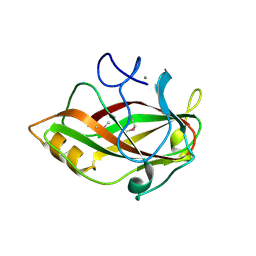 | | STRUCTURE OF FAM17 CARBOHYDRATE BINDING MODULE FROM CLOSTRIDIUM CELLULOVORANS | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA GLUCANASE ENGF | | Authors: | Notenboom, V, Boraston, A.B, Chiu, P, Freelove, A.C.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-05-20 | | Release date: | 2001-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition of cello-oligosaccharides by a family 17 carbohydrate-binding module: an X-ray crystallographic, thermodynamic and mutagenic study.
J.Mol.Biol., 314, 2001
|
|
1DR1
 
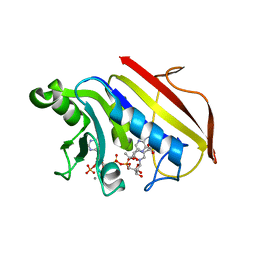 | | 2.2 ANGSTROMS CRYSTAL STRUCTURE OF CHICKEN LIVER DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADP+ AND BIOPTERIN | | Descriptor: | 7,8-DIHYDROBIOPTERIN, CALCIUM ION, DIHYDROFOLATE REDUCTASE, ... | | Authors: | Mctigue, M.A, Davies /II, J.F, Kaufman, B.T, Xuong, N.-H, Kraut, J. | | Deposit date: | 1992-03-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of chicken liver dihydrofolate reductase complexed with NADP+ and biopterin.
Biochemistry, 31, 1992
|
|
2HB9
 
 | | Crystal Structure of the Zinc-Beta-Lactamase L1 from Stenotrophomonas Maltophilia (Inhibitor 3) | | Descriptor: | 4-AMINO-5-(2-METHYLPHENYL)-2,4-DIHYDRO-3H-1,2,4-TRIAZOLE-3-THIONE, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Nauton, L, Garau, G, Kahn, R, Dideberg, O. | | Deposit date: | 2006-06-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the design of inhibitors for the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia.
J.Mol.Biol., 375, 2008
|
|
1DAJ
 
 | | COMPARISON OF TERNARY COMPLEXES OF PNEUMOCYSTIS CARINII AND WILD TYPE HUMAN DIHYDROFOLATE REDUCTASE WITH COENZYME NADPH AND A NOVEL CLASSICAL ANTITUMOR FURO[2,3D]PYRIMIDINE ANTIFOLATE | | Descriptor: | DIHYDROFOLATE REDUCTASE, N-[4-[(2,4-DIAMINOFURO[2,3D]PYRIMIDIN-5-YL)METHYL]METHYLAMINO]-BENZOYL]-L-GLUTAMATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Gangjee, A, Devraj, R, Queener, S.F, Blakley, R.L. | | Deposit date: | 1997-07-29 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comparison of ternary complexes of Pneumocystis carinii and wild-type human dihydrofolate reductase with coenzyme NADPH and a novel classical antitumor furo[2,3-d]pyrimidine antifolate.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1W1Z
 
 | | Structure of the plant like 5-Aminolaevulinic Acid Dehydratase from Chlorobium vibrioforme | | Descriptor: | DELTA-AMINOLEVULINIC ACID DEHYDRATASE, LAEVULINIC ACID, MAGNESIUM ION | | Authors: | Coates, L, Beaven, G, Erskine, P.T, Beale, S.I, Avissar, Y.J, Gill, R, Mohammed, F, Wood, S.P, Shoolingin-Jordan, P, Cooper, J.B. | | Deposit date: | 2004-06-24 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The X-ray structure of the plant like 5-aminolaevulinic acid dehydratase from Chlorobium vibrioforme complexed with the inhibitor laevulinic acid at 2.6 A resolution.
J. Mol. Biol., 342, 2004
|
|
2FPB
 
 | |
2FP8
 
 | |
2FPC
 
 | |
2LDX
 
 | |
2FQ1
 
 | | Crystal structure of the two-domain non-ribosomal peptide synthetase EntB containing isochorismate lyase and aryl-carrier protein domains | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Isochorismatase, ... | | Authors: | Drake, E.J, Nicolai, D.A, Gulick, A.M. | | Deposit date: | 2006-01-17 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the EntB multidomain nonribosomal peptide synthetase and functional analysis of its interaction with the EntE adenylation domain.
Chem.Biol., 13, 2006
|
|
2FP9
 
 | | Crystal structure of Native Strictosidine Synthase | | Descriptor: | L(+)-TARTARIC ACID, Strictosidine synthase | | Authors: | Panjikar, S. | | Deposit date: | 2006-01-16 | | Release date: | 2006-05-23 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The structure of Rauvolfia serpentina strictosidine synthase is a novel six-bladed beta-propeller fold in plant proteins
Plant Cell, 18, 2006
|
|
2LYM
 
 | |
