3NOV
 
 | | Crystal Structure of D17E Isocyanide Hydratase from Pseudomonas fluorescens | | Descriptor: | ACETATE ION, ThiJ/PfpI family protein | | Authors: | Lakshminarasimhan, M, Madzelan, P, Nan, R, Milkovic, N.M, Wilson, M.A. | | Deposit date: | 2010-06-25 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Evolution of New Enzymatic Function by Structural Modulation of Cysteine Reactivity in Pseudomonas fluorescens Isocyanide Hydratase.
J.Biol.Chem., 285, 2010
|
|
3NOO
 
 | | Crystal Structure of C101A Isocyanide Hydratase from Pseudomonas fluorescens | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ThiJ/PfpI family protein | | Authors: | Lakshminarasimhan, M, Madzelan, P, Nan, R, Milkovic, N.M, Wilson, M.A. | | Deposit date: | 2010-06-25 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Evolution of New Enzymatic Function by Structural Modulation of Cysteine Reactivity in Pseudomonas fluorescens Isocyanide Hydratase.
J.Biol.Chem., 285, 2010
|
|
5MAJ
 
 | | CATHEPSIN L IN COMPLEX WITH 4-[cyclopentyl(imidazo[1,2-a]pyridin-2-ylmethyl)amino]-6-morpholino-1,3,5-triazine-2-carbonitrile | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L1, ~{N}-cyclopentyl-~{N}-(imidazo[1,2-a]pyridin-2-ylmethyl)-4-(iminomethyl)-6-morpholin-4-yl-1,3,5-triazin-2-amine | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Inhibition of the Cysteine Protease Human Cathepsin L by Triazine Nitriles: AmideHeteroarene pi-Stacking Interactions and Chalcogen Bonding in the S3 Pocket.
ChemMedChem, 12, 2017
|
|
6HGY
 
 | | CRYSTAL STRUCTURE OF CATHEPSIN K WITH N-DESMETHYL THALASSOSPIRAMIDE C | | Descriptor: | Cathepsin K, THALASSOSPIRAMIDE C | | Authors: | Zakarian, A, Buckman, B.O, Adler, M, Griessner, A, Blaesse, M. | | Deposit date: | 2018-08-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Total Synthesis of Covalent Cysteine Protease Inhibitor N-Desmethyl Thalassospiramide C and Crystallographic Evidence for Its Mode of Action.
Org.Lett., 21, 2019
|
|
5MAE
 
 | | CATHEPSIN L IN COMPLEX WITH (2S,4R)-4-(2-Chloro-4-methoxy-benzenesulfonyl)-1-[3-(5-chloro-pyridin-2-yl)-azetidine-3-carbonyl]-pyrrolidine-2-car boxylic acid (1-cyano-cyclopropyl)-amide | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L1, [4-[cyclopentyl(pyrazin-2-ylmethyl)amino]-6-morpholin-4-yl-1,3,5-triazin-2-yl]methylideneazanide | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Inhibition of the Cysteine Protease Human Cathepsin L by Triazine Nitriles: AmideHeteroarene pi-Stacking Interactions and Chalcogen Bonding in the S3 Pocket.
ChemMedChem, 12, 2017
|
|
3UEY
 
 | |
6B2M
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase in complex with coenzyme A | | Descriptor: | ATP-utilizing enzyme of the PP-loopsuperfamily, COENZYME A, PHOSPHATE ION | | Authors: | Fellner, M, Hausinger, R.P, Hu, J. | | Deposit date: | 2017-09-20 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Analysis of the Active Site Cysteine Residue of the Sacrificial Sulfur Insertase LarE from Lactobacillus plantarum.
Biochemistry, 57, 2018
|
|
3UEJ
 
 | |
3W55
 
 | | The structure of ERK2 in complex with FR148083 | | Descriptor: | (3S,5Z,8S,9S,11E)-8,9,16-trihydroxy-14-methoxy-3-methyl-3,4,9,10-tetrahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Mitogen-activated protein kinase 1 | | Authors: | Ohori, M, Kinoshita, T. | | Deposit date: | 2013-01-21 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Role of a cysteine residue in the active site of ERK and the MAPKK family
Biochem.Biophys.Res.Commun., 353, 2007
|
|
5VSL
 
 | | Crystal structure of viperin with bound [4Fe-4S] cluster and S-adenosylhomocysteine (SAH) | | Descriptor: | IRON/SULFUR CLUSTER, Radical S-adenosyl methionine domain-containing protein 2, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Fenwick, M.K, Li, Y, Cresswell, P, Modis, Y, Ealick, S.E. | | Deposit date: | 2017-05-11 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Structural studies of viperin, an antiviral radical SAM enzyme.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1JF9
 
 | |
2VU2
 
 | | Biosynthetic thiolase from Z. ramigera. Complex with S-pantetheine-11- pivalate. | | Descriptor: | (3R)-3-hydroxy-2,2-dimethyl-4-oxo-4-({3-oxo-3-[(2-sulfanylethyl)amino]propyl}amino)butyl 2,2-dimethylpropanoate, ACETYL-COA ACETYLTRANSFERASE, SULFATE ION | | Authors: | Kursula, P, Merilainen, G, Schmitz, W, Wierenga, R.K. | | Deposit date: | 2008-05-19 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Sulfur Atoms of the Substrate Coa and the Catalytic Cysteine are Required for a Productive Mode of Substrate Binding in Bacterial Biosynthetic Thiolase, a Thioester-Dependent Enzyme.
FEBS J., 275, 2008
|
|
3P7X
 
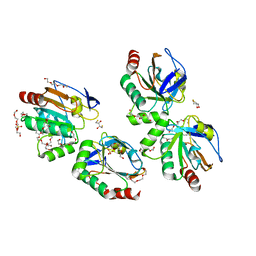 | | Crystal structure of an atypical two-cysteine peroxiredoxin (SAOUHSC_01822) from Staphylococcus aureus NCTC8325 | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Probable thiol peroxidase, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2010-10-13 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of an atypical two-cysteine peroxiredoxin (SAOUHSC_01822) from Staphylococcus aureus NCTC8325
To be Published
|
|
8BVC
 
 | | Solution structure of Metridium senile toxin Ms13-1 with the unique fold | | Descriptor: | Ms13-1 | | Authors: | Mineev, K.S, Arseniev, A.S, Andreev, Y.A, Osmakov, D.I, Khasanov, T.A. | | Deposit date: | 2022-12-06 | | Release date: | 2023-12-20 | | Last modified: | 2025-07-09 | | Method: | SOLUTION NMR | | Cite: | Sea anemone Cys-ladder peptide Ms13-1 induces a pain response as a positive modulator of acid-sensing ion channel 1a.
Febs J., 292, 2025
|
|
5LQ6
 
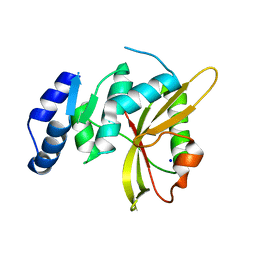 | | Salmonella effector SpvD - R161 variant | | Descriptor: | SODIUM ION, Virulence protein vsdE | | Authors: | Zhang, Y, Grabe, G.J, Rolhion, N, Yang, Y, Holden, D.W, Hare, S.A. | | Deposit date: | 2016-08-16 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The Salmonella Effector SpvD Is a Cysteine Hydrolase with a Serovar-specific Polymorphism Influencing Catalytic Activity, Suppression of Immune Responses, and Bacterial Virulence.
J. Biol. Chem., 291, 2016
|
|
5LQ7
 
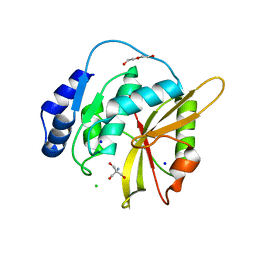 | | Salmonella effector SpvD - G161 variant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Przydacz, M, Grabe, G.J, Holden, D.W, Hare, S.A. | | Deposit date: | 2016-08-16 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Salmonella Effector SpvD Is a Cysteine Hydrolase with a Serovar-specific Polymorphism Influencing Catalytic Activity, Suppression of Immune Responses, and Bacterial Virulence.
J. Biol. Chem., 291, 2016
|
|
5CGH
 
 | |
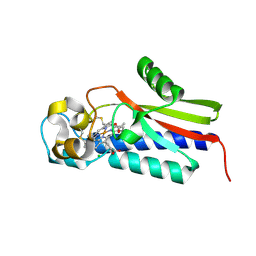
5B82
 
 | |
1LDR
 
 | |
6J76
 
 | | Structure of 3,6-anhydro-L-galactose Dehydrogenase in Complex with NAP | | Descriptor: | Aldehyde dehydrogenase A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, P.Y, Wang, Y, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2019-01-17 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.368 Å) | | Cite: | 3,6-Anhydro-L-Galactose Dehydrogenase VvAHGD is a Member of a New Aldehyde Dehydrogenase Family and Catalyzes by a Novel Mechanism with Conformational Switch of Two Catalytic Residues Cysteine 282 and Glutamate 248.
J.Mol.Biol., 432, 2020
|
|
5LAC
 
 | |
6J75
 
 | | Structure of 3,6-anhydro-L-galactose Dehydrogenase | | Descriptor: | Aldehyde dehydrogenase A | | Authors: | Li, P.Y, Wang, Y, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2019-01-17 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | 3,6-Anhydro-L-Galactose Dehydrogenase VvAHGD is a Member of a New Aldehyde Dehydrogenase Family and Catalyzes by a Novel Mechanism with Conformational Switch of Two Catalytic Residues Cysteine 282 and Glutamate 248.
J.Mol.Biol., 432, 2020
|
|
207L
 
 | | MUTANT HUMAN LYSOZYME C77A | | Descriptor: | LYSOZYME, N-ACETYL-L-CYSTEINE | | Authors: | Matsushima, M, Song, H. | | Deposit date: | 1996-03-26 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A role of PDI in the reductive cleavage of mixed disulfides.
J.Biochem.(Tokyo), 120, 1996
|
|
1XDJ
 
 | |
1XU1
 
 | | The crystal structure of APRIL bound to TACI | | Descriptor: | NICKEL (II) ION, Tumor necrosis factor ligand superfamily member 13, Tumor necrosis factor receptor superfamily member 13B | | Authors: | Hymowitz, S.G, Patel, D.R, Wallweber, H.J.A, Runyon, S, Yan, M, Yin, J, Shriver, S.K, Gordon, N.C, Pan, B, Skelton, N.J, Kelley, R.F, Starovasnik, M.A. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of APRIL-receptor complexes: Like BCMA, TACI employs only a single cysteine-rich domain for high-affinity ligand binding
J.Biol.Chem., 280, 2005
|
|
