5E3J
 
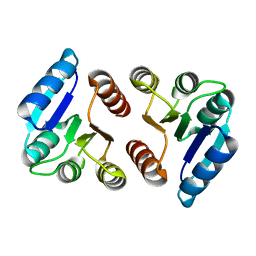 | | The response regulator RstA is a potential drug target for Acinetobacter baumannii | | Descriptor: | Response regulator RstA | | Authors: | Russo, T.A, Manohar, A, Beanan, J.M, Olson, R, MacDonald, U, Graham, J, Umland, T.C. | | Deposit date: | 2015-10-02 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Response Regulator BfmR Is a Potential Drug Target for Acinetobacter baumannii.
Msphere, 1, 2016
|
|
5E5C
 
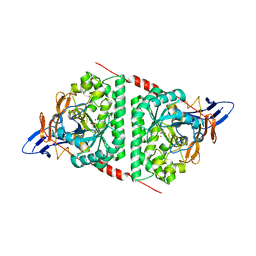 | | Crystal structure of dihydropyrimidinase from Pseudomonas aeruginosa PAO1 | | Descriptor: | D-hydantoinase/dihydropyrimidinase, ZINC ION | | Authors: | Huang, C.C, Huang, Y.H, Hsieh, Y.C, Tzeng, C.T, Chen, C.J, Huang, C.Y. | | Deposit date: | 2015-10-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of dihydropyrimidinase from Pseudomonas aeruginosa PAO1: Insights into the molecular basis of formation of a dimer
Biochem.Biophys.Res.Commun., 478, 2016
|
|
1WVA
 
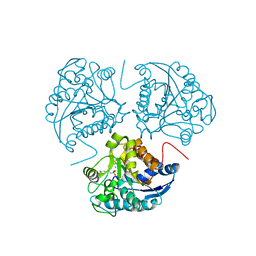 | | Crystal structure of human arginase I from twinned crystal | | Descriptor: | Arginase 1, MANGANESE (II) ION, S-2-(BORONOETHYL)-L-CYSTEINE | | Authors: | Di Costanzo, L, Sabio, G, Mora, A, Rodriguez, P.C, Ochoa, A.C, Centeno, F, Christianson, D.W. | | Deposit date: | 2004-12-14 | | Release date: | 2005-09-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of human arginase I at 1.29 A resolution and exploration of inhibition in the immune response
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3E8Q
 
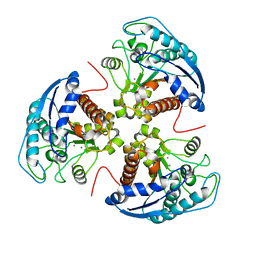 | | X-ray structure of rat arginase I-T135A: the unliganded complex | | Descriptor: | Arginase-1, MANGANESE (II) ION | | Authors: | Shishova, E.Y, Di Costanzo, L, Emig, F.A, Ash, D.E, Christianson, D.W. | | Deposit date: | 2008-08-20 | | Release date: | 2008-12-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Probing the specificity determinants of amino acid recognition by arginase.
Biochemistry, 48, 2009
|
|
5XQH
 
 | | Crystal structure of truncated human Rogdi | | Descriptor: | Protein rogdi homolog | | Authors: | Lee, H, Lee, C. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The crystal structure of human Rogdi provides insight into the causes of Kohlschutter-Tonz Syndrome
Sci Rep, 7, 2017
|
|
3E6K
 
 | |
6EQA
 
 | | HLA class I histocompatibility antigen | | Descriptor: | ALA-ALA-GLY-ILE-GLY-ILE-LEU-THR-VAL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Rizkallah, P.J, Cole, D.K. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | TCR-induced alteration of primary MHC peptide anchor residue.
Eur.J.Immunol., 49, 2019
|
|
2D3I
 
 | | Crystal Structure of Aluminum-Bound Ovotransferrin at 2.15 Angstrom Resolution | | Descriptor: | ALUMINUM ION, BICARBONATE ION, Ovotransferrin | | Authors: | Mizutani, K, Mikami, B, Aibara, S, Hirose, M. | | Deposit date: | 2005-09-28 | | Release date: | 2005-11-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of aluminium-bound ovotransferrin at 2.15 Angstroms resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5E9E
 
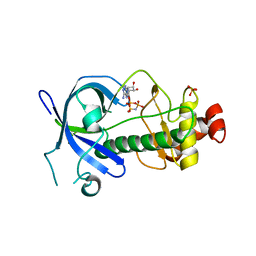 | |
3VXW
 
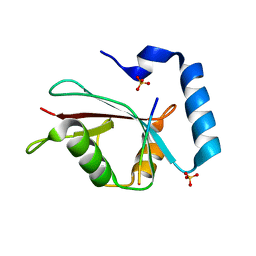 | |
6EAZ
 
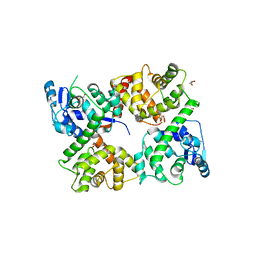 | |
3VP3
 
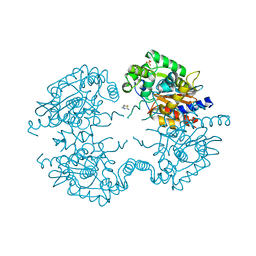 | | Crystal structure of human glutaminase in complex with inhibitor 3 | | Descriptor: | 5,5'-pentane-1,5-diylbis(1,3,4-thiadiazol-2-amine), Glutaminase kidney isoform, mitochondrial, ... | | Authors: | Thangavelu, K, Sivaraman, J. | | Deposit date: | 2012-02-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VPH
 
 | | L-lactate dehydrogenase from Thermus caldophilus GK24 complexed with oxamate, NADH and FBP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, L-lactate dehydrogenase, ... | | Authors: | Arai, K, Ohno, T, Miyanaga, A, Fushinobu, S, Taguchi, H. | | Deposit date: | 2012-03-01 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The core of allosteric motion in Thermus caldophilus L-lactate dehydrogenase.
J.Biol.Chem., 2014
|
|
5A4R
 
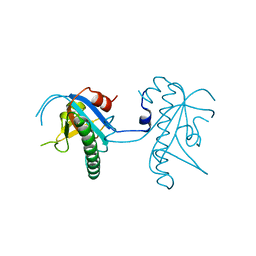 | | Crystal structure of a vitamin B12 trafficking protein | | Descriptor: | METHYLMALONIC ACIDURIA AND HOMOCYSTINURIA TYPE D HOMOLOG, MITOCHONDRIAL | | Authors: | Kopec, J, Fitzpatrick, F, Froese, D.S, Velupillai, S, Nowak, R, Chalk, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Fowler, B, Baumgartner, M.R, Yue, W.W. | | Deposit date: | 2015-06-11 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insights Into the Mmachc-Mmadhc Protein Complex Involved in Vitamin B12 Trafficking.
J.Biol.Chem., 290, 2015
|
|
6EH6
 
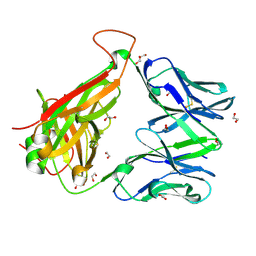 | |
1I8L
 
 | | HUMAN B7-1/CTLA-4 CO-STIMULATORY COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTOTOXIC T-LYMPHOCYTE PROTEIN 4, T LYMPHOCYTE ACTIVATION ANTIGEN CD80, ... | | Authors: | Stamper, C.C, Somers, W.S, Mosyak, L. | | Deposit date: | 2001-03-14 | | Release date: | 2001-04-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the B7-1/CTLA-4 complex that inhibits human immune responses.
Nature, 410, 2001
|
|
2YP5
 
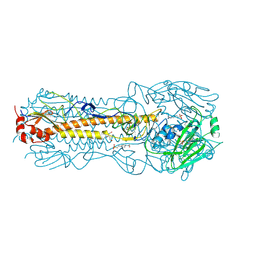 | | Haemagglutinin of 2004 Human H3N2 Virus in Complex with Avian Receptor Analogue 3SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | Deposit date: | 2012-10-29 | | Release date: | 2012-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5DYJ
 
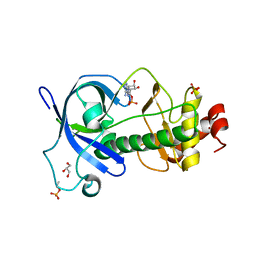 | | Mysosin heavy chain kinase A catalytic domain mutant - D663A | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Myosin heavy chain kinase A, ... | | Authors: | van Staalduinen, L.M, Yang, Y, Jia, Z. | | Deposit date: | 2015-09-24 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Dictyostelium Myosin-II Heavy Chain Kinase A (MHCK-A) alpha-kinase domain apoenzyme reveals a novel autoinhibited conformation.
Sci Rep, 6, 2016
|
|
5DZW
 
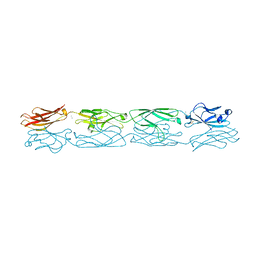 | | Protocadherin alpha 4 extracellular cadherin domains 1-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin alpha-4, ... | | Authors: | Goodman, K.M, Bahna, F, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2015-09-26 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural Basis of Diverse Homophilic Recognition by Clustered alpha- and beta-Protocadherins.
Neuron, 90, 2016
|
|
5XTC
 
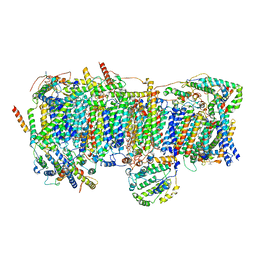 | | Cryo-EM structure of human respiratory complex I transmembrane arm | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, Acyl carrier protein, ... | | Authors: | Gu, J, Wu, M, Yang, M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-08-30 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Architecture of Human Mitochondrial Respiratory Megacomplex I2III2IV2.
Cell, 170, 2017
|
|
6EPA
 
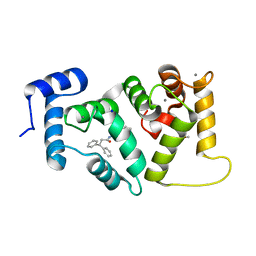 | | Structure of dNCS-1 bound to the NCS-1/Ric8a protein/protein interaction regulator IGS-1.76 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, FI18190p1, ... | | Authors: | Sanchez-Barrena, M.J, Daniel, M, Infantes, L. | | Deposit date: | 2017-10-11 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Deciphering the Inhibition of the Neuronal Calcium Sensor 1 and the Guanine Exchange Factor Ric8a with a Small Phenothiazine Molecule for the Rational Generation of Therapeutic Synapse Function Regulators.
J. Med. Chem., 61, 2018
|
|
6ES1
 
 | |
5ELR
 
 | |
6ERZ
 
 | | The crystal structure of mouse chloride intracellular channel protein 6 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Chloride intracellular channel protein 6, SULFATE ION | | Authors: | Ferofontov, A, Giladi, M, Haitin, Y. | | Deposit date: | 2017-10-19 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Inherent flexibility of CLIC6 revealed by crystallographic and solution studies.
Sci Rep, 8, 2018
|
|
3VP1
 
 | |
