8IHI
 
 | | Cryo-EM structure of HCA2-Gi complex with acifran | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHF
 
 | | Cryo-EM structure of HCA2-Gi complex with MK6892 | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHJ
 
 | | Cryo-EM structure of HCA3-Gi complex with acifran | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IC8
 
 | | Exo-alpha-D-arabinofuranosidase from Microbacterium arabinogalactanolyticum | | Descriptor: | Exo-alpha-D-arabinofuranosidase, PHOSPHATE ION | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
8IHK
 
 | | Cryo-EM structure of HCA3-Gi complex with acifran (local) | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, Soluble cytochrome b562,Hydroxycarboxylic acid receptor 3 | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHH
 
 | | Cryo-EM structure of HCA2-Gi complex with LUF6283 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-butyl-1~{H}-pyrazole-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8HZZ
 
 | | GuApiGT (UGT79B74) | | Descriptor: | SULFATE ION, apiosyltransferase | | Authors: | Wang, H.T, Wang, Z.L, Li, F.D, Ye, M. | | Deposit date: | 2023-01-10 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into the missing apiosylation step in flavonoid apiosides biosynthesis of Leguminosae plants.
Nat Commun, 14, 2023
|
|
8I0E
 
 | | Sb3GT1 complex with UDP | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, H.T, Wang, Z.L, Ye, M. | | Deposit date: | 2023-01-10 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the missing apiosylation step in flavonoid apiosides biosynthesis of Leguminosae plants.
Nat Commun, 14, 2023
|
|
6E7C
 
 | | 14-pf 3-start GMPCPP-human alpha1B/beta2B microtubules | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Ti, S.C, Alushin, G.M, Kapoor, T.M. | | Deposit date: | 2018-07-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Human beta-Tubulin Isotypes Can Regulate Microtubule Protofilament Number and Stability.
Dev. Cell, 47, 2018
|
|
8CDQ
 
 | | Plasmodium falciparum Myosin A full-length, post-rigor state complexed to the inhibitor KNX-002 and Mg.ATP-gamma-S | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-methoxyphenyl)-~{N}-[(3-thiophen-2-yl-1~{H}-pyrazol-4-yl)methyl]cyclopropan-1-amine, GLYCEROL, ... | | Authors: | Moussaoui, D, Robblee, J.P, Robert-Paganin, J, Auguin, D, Fisher, F, Fagnant, P.M, MacFarlane, J.E, Schaletzky, J, Wehri, E, Mueller-Dieckmann, C, Baum, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2023-01-31 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mechanism of small molecule inhibition of Plasmodium falciparum myosin A informs antimalarial drug design.
Nat Commun, 14, 2023
|
|
8C76
 
 | | Light-state 2.5 Angstrom wild-type X-ray crystal structure of the cobalamin binding domain belonging to a light-dependent transcription regulator TtCarH obtained under aerobic conditions from a form 2 crystal illuminated during 5 s | | Descriptor: | COBALAMIN, Probable transcriptional regulator | | Authors: | Rios-Santacruz, R, Colletier, J.P, Schiro, G, Weik, M. | | Deposit date: | 2023-01-12 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Redox driven B 12 -ligand switch drives CarH photoresponse.
Nat Commun, 14, 2023
|
|
8C73
 
 | | Dark state 1.8 Angstrom crystal structure of cobalamin binding domain belonging to a light-dependent transcription regulator TtCarH obtained under aerobic condition form ll | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Probable transcriptional regulator | | Authors: | Rios-Santacruz, R, Colletier, J.P, Schiro, G, Weik, M. | | Deposit date: | 2023-01-12 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Redox driven B 12 -ligand switch drives CarH photoresponse.
Nat Commun, 14, 2023
|
|
8CEA
 
 | | Cytochrome c maturation complex CcmABCD, E154Q | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, Heme exporter protein C, ... | | Authors: | Ilcu, L, Zhang, L, Einsle, O. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Architecture of the Heme-translocating CcmABCD/E complex required for Cytochrome c maturation.
Nat Commun, 14, 2023
|
|
8C9G
 
 | | Priestia megaterium mupirocin-sensitive isoleucyl-tRNA synthetase 1 complexed with mupirocin | | Descriptor: | CHLORIDE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | Authors: | Brkic, A, Leibundgut, M, Jablonska, J, Zanki, V, Car, Z, Petrovic Perokovic, V, Ban, N, Gruic-Sovulj, I. | | Deposit date: | 2023-01-22 | | Release date: | 2023-08-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Nat Commun, 14, 2023
|
|
8C8U
 
 | | Priestia megaterium mupirocin-resistant isoleucyl-tRNA synthetase 2 complexed with mupirocin | | Descriptor: | Isoleucine--tRNA ligase, L(+)-TARTARIC ACID, MUPIROCIN, ... | | Authors: | Brkic, A, Leibundgut, M, Jablonska, J, Zanki, V, Car, Z, Petrovic Perokovic, V, Gruic-Sovulj, I, Ban, N. | | Deposit date: | 2023-01-21 | | Release date: | 2023-08-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Nat Commun, 14, 2023
|
|
8CE1
 
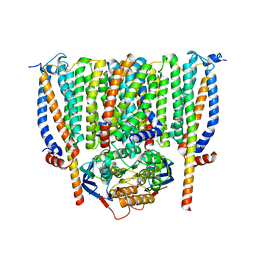 | | Cytochrome c maturation complex CcmABCD | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, Heme exporter protein C, ... | | Authors: | Ilcu, L, Zhang, L, Einsle, O. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Architecture of the Heme-translocating CcmABCD/E complex required for Cytochrome c maturation.
Nat Commun, 14, 2023
|
|
8C8W
 
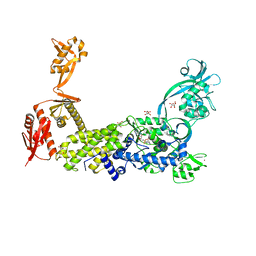 | | Priestia megaterium mupirocin hyper-resistant HIGH motif mutant of type 2 isoleucyl-tRNA synthetase complexed with an isoleucyl-adenylate analogue | | Descriptor: | D(-)-TARTARIC ACID, Isoleucine--tRNA ligase, N-[ISOLEUCINYL]-N'-[ADENOSYL]-DIAMINOSUFONE, ... | | Authors: | Brkic, A, Leibundgut, M, Jablonska, J, Zanki, V, Car, Z, Petrovic Perokovic, V, Ban, N, Gruic-Sovulj, I. | | Deposit date: | 2023-01-21 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Nat Commun, 14, 2023
|
|
9EZ1
 
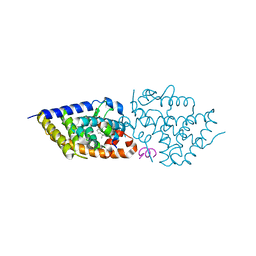 | | Vitamin D receptor in complex with 1,4a,25-trihydroxyvitamin D3 | | Descriptor: | 1,4a,25-trihydroxyvitamin D3, ACETATE ION, Nuclear receptor coactivator 2, ... | | Authors: | Rochel, N. | | Deposit date: | 2024-04-10 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 4-Hydroxy-1 alpha ,25-Dihydroxyvitamin D 3 : Synthesis and Structure-Function Study.
Biomolecules, 14, 2024
|
|
8C9F
 
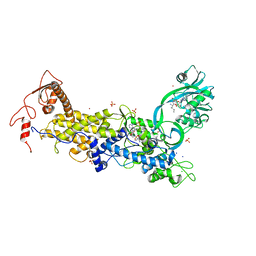 | | Priestia megaterium inactive HIGH motif mutant of type1 isoleucyl-tRNA synthetase complexed with an isoleucyl-adenylate analogue. | | Descriptor: | CHLORIDE ION, Isoleucine--tRNA ligase, LITHIUM ION, ... | | Authors: | Brkic, A, Leibundgut, M, Jablonska, J, Zanki, V, Car, Z, Petrovic Perokovic, V, Ban, N, Gruic-Sovulj, I. | | Deposit date: | 2023-01-21 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Nat Commun, 14, 2023
|
|
8C9D
 
 | | Priestia megaterium W130Q mutant of type 2 isoleucyl-tRNA synthetase complexed with an isoleucyl-adenylate analogue | | Descriptor: | D(-)-TARTARIC ACID, Isoleucine--tRNA ligase, N-[ISOLEUCINYL]-N'-[ADENOSYL]-DIAMINOSUFONE, ... | | Authors: | Brkic, A, Leibundgut, M, Jablonska, J, Zanki, V, Car, Z, Petrovic Perokovic, V, Ban, N, Gruic-Sovulj, I. | | Deposit date: | 2023-01-21 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Nat Commun, 14, 2023
|
|
8C36
 
 | |
8C32
 
 | |
8C8V
 
 | | Priestia megaterium mupirocin-resistant isoleucyl-tRNA synthetase 2 with a fully-resolved C-terminal tRNA-binding domain complexed with an isoleucyl-adenylate analogue | | Descriptor: | D(-)-TARTARIC ACID, Isoleucine--tRNA ligase, N-[ISOLEUCINYL]-N'-[ADENOSYL]-DIAMINOSUFONE, ... | | Authors: | Brkic, A, Leibundgut, M, Jablonska, J, Zanki, V, Car, Z, Petrovic Perokovic, V, Ban, N, Gruic-Sovulj, I. | | Deposit date: | 2023-01-21 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Nat Commun, 14, 2023
|
|
8C33
 
 | |
8C35
 
 | |
