1XAT
 
 | |
1LKP
 
 | | Crystal structure of Desulfovibrio vulgaris rubrerythrin all-iron(II) form, azide adduct | | Descriptor: | AZIDE ION, FE (II) ION, Rubrerythrin | | Authors: | Jin, S, Kurtz Jr, D.M, Liu, Z.J, Rose, J, Wang, B.C. | | Deposit date: | 2002-04-25 | | Release date: | 2002-09-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | X-ray Crystal Structures of Reduced Rubrerythrin and its Azide Adduct: A Structure-Based Mechanism for a Non-Heme Diiron Peroxidase
J.Am.Chem.Soc., 124, 2002
|
|
1L5Q
 
 | | Human liver glycogen phosphorylase a complexed with caffeine, N-Acetyl-beta-D-glucopyranosylamine, and CP-403700 | | Descriptor: | CAFFEINE, Glycogen phosphorylase, liver form, ... | | Authors: | Ekstrom, J.L, Pauly, T.A, Carty, M.D, Soeller, W.C, Culp, J, Danley, D.E, Hoover, D.J, Treadway, J.L, Gibbs, E.M, Fletterick, R.J, Day, Y.S.N, Myszka, D.G, Rath, V.L. | | Deposit date: | 2002-03-07 | | Release date: | 2002-12-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-activity analysis of the purine binding
site of human liver glycogen phosphorylase.
Chem.Biol., 9, 2002
|
|
1LQX
 
 | | Crystal structure of V45E mutant of cytochrome b5 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome b5 | | Authors: | Gan, J.-H, Wu, J, Wang, Z.-Q, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2002-05-14 | | Release date: | 2002-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of V45E and V45Y mutants and structure comparison of a variety of cytochrome b5 mutants.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1LT3
 
 | | HEAT-LABILE ENTEROTOXIN DOUBLE MUTANT N40C/G166C | | Descriptor: | HEAT-LABILE ENTEROTOXIN, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Van Den Akker, F, Hol, W.G.J. | | Deposit date: | 1997-04-12 | | Release date: | 1997-07-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of heat-labile enterotoxin from Escherichia coli with increased thermostability introduced by an engineered disulfide bond in the A subunit.
Protein Sci., 6, 1997
|
|
1LBS
 
 | | LIPASE (E.C.3.1.1.3) (TRIACYLGLYCEROL HYDROLASE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LIPASE B, N-HEXYLPHOSPHONATE ETHYL ESTER | | Authors: | Uppenberg, J, Jones, T.A. | | Deposit date: | 1995-07-11 | | Release date: | 1995-10-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic and molecular-modeling studies of lipase B from Candida antarctica reveal a stereospecificity pocket for secondary alcohols.
Biochemistry, 34, 1995
|
|
2ACQ
 
 | | AN ANION BINDING SITE IN HUMAN ALDOSE REDUCTASE: MECHANISTIC IMPLICATIONS FOR THE BINDING OF CITRATE, CACODYLATE, AND GLUCOSE-6-PHOSPHATE | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H, Bohren, K.M, Gabbay, K.H, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-04-15 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An anion binding site in human aldose reductase: mechanistic implications for the binding of citrate, cacodylate, and glucose 6-phosphate.
Biochemistry, 33, 1994
|
|
1LIN
 
 | | CALMODULIN COMPLEXED WITH TRIFLUOPERAZINE (1:4 COMPLEX) | | Descriptor: | 10-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPYL]-2-TRIFLUOROMETHYL-10H-PHENOTHIAZINE, CALCIUM ION, CALMODULIN | | Authors: | Vandonselaar, M, Hickie, R.A, Quail, J.W, Delbaere, L.T.J. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trifluoperazine-induced conformational change in Ca(2+)-calmodulin.
Nat.Struct.Biol., 1, 1994
|
|
1LDI
 
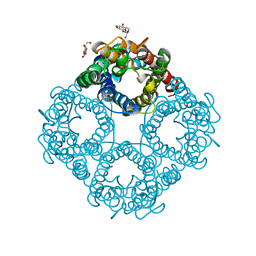 | | CRYSTAL STRUCTURE OF THE E. COLI GLYCEROL FACILITATOR (GLPF) WITHOUT SUBSTRATE GLYCEROL | | Descriptor: | Glycerol uptake facilitator protein, octyl beta-D-glucopyranoside | | Authors: | Nollert, P, Miercke, L.J.W, O'Connell, J, Stroud, R.M. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Control of the selectivity of the aquaporin water channel family by global orientational tuning.
Science, 296, 2002
|
|
1LCE
 
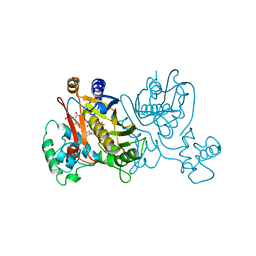 | | LACTOBACILLUS CASEI THYMIDYLATE SYNTHASE TERNARY COMPLEX WITH DUMP AND CH2THF | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5,10-METHYLENE-6-HYDROFOLIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Birdsall, D.L, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1995-06-22 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refined structures of substrate-bound and phosphate-bound thymidylate synthase from Lactobacillus casei.
J.Mol.Biol., 232, 1993
|
|
1LOA
 
 | |
2A1N
 
 | |
1LJU
 
 | | X-RAY STRUCTURE OF C15A ARSENATE REDUCTASE FROM PI258 COMPLEXED WITH ARSENITE | | Descriptor: | POTASSIUM ION, arsenate reductase | | Authors: | Zegers, I, Martins, J.C, Willem, R, Wyns, L, Messens, J. | | Deposit date: | 2002-04-22 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | All intermediates of the arsenate reductase mechanism, including an intramolecular dynamic disulfide cascade.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LSJ
 
 | |
1LT4
 
 | | HEAT-LABILE ENTEROTOXIN MUTANT S63K | | Descriptor: | HEAT-LABILE ENTEROTOXIN, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Van Den Akker, F, Hol, W.G.J. | | Deposit date: | 1997-04-14 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a non-toxic mutant of heat-labile enterotoxin, which is a potent mucosal adjuvant.
Protein Sci., 6, 1997
|
|
1LTW
 
 | | RECOMBINANT SPERM WHALE MYOGLOBIN 29W MUTANT (OXY) | | Descriptor: | MYOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Li, T, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1996-11-02 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Perturbation of the Fe-O2 Bond by Nearby Residues in Heme Pocket: Observation of Vfe-O2 Raman Bands for Oxymyoglobin Mutants
J.Am.Chem.Soc., 118, 1996
|
|
2AC2
 
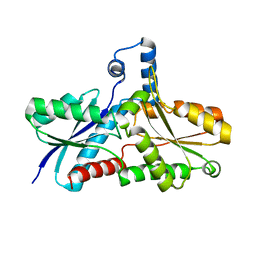 | | Crystal structure of the Tyr13Phe mutant variant of Bacillus subtilis Ferrochelatase with Zn(2+) bound at the active site | | Descriptor: | Ferrochelatase, ZINC ION | | Authors: | Shipovskov, S, Karlberg, T, Fodje, M, Hansson, M.D, Ferreira, G.C, Hansson, M, Reimann, C.T, Al-Karadaghi, S. | | Deposit date: | 2005-07-18 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Metallation of the Transition-state Inhibitor N-methyl Mesoporphyrin by Ferrochelatase: Implications for the Catalytic Reaction Mechanism.
J.Mol.Biol., 352, 2005
|
|
2ACA
 
 | | X-ray structure of a putative adenylate cyclase Q87NV8 from Vibrio parahaemolyticus at the 2.25 A resolution. Northeast Structural Genomics Target VpR19. | | Descriptor: | PHOSPHATE ION, putative adenylate cyclase | | Authors: | Kuzin, A.P, Abashidze, M, Vorobiev, S.M, Forouhar, F, Chen, Y, Acton, T, Xiao, R, Conover, K, Ma, L.-C, Cunningham, K.E, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-07-18 | | Release date: | 2005-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: |
|
|
1LM5
 
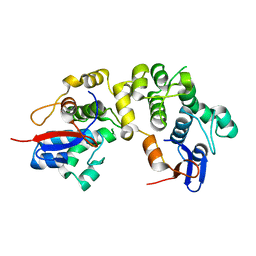 | | Structures of two intermediate filament-binding fragments of desmoplakin reveal a unique repeat motif structure | | Descriptor: | subdomain of Desmoplakin Carboxy-Terminal domain (DPCT) | | Authors: | Choi, H.J, Park-Snyder, S, Pascoe, L.T, Green, K.J, Weis, W.I. | | Deposit date: | 2002-04-30 | | Release date: | 2002-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of two intermediate filament-binding fragments of desmoplakin reveal a unique repeat motif structure.
Nat.Struct.Biol., 9, 2002
|
|
1LPB
 
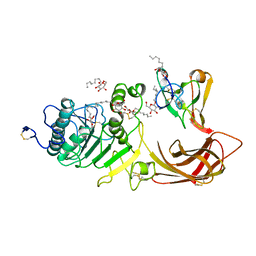 | | THE 2.46 ANGSTROMS RESOLUTION STRUCTURE OF THE PANCREATIC LIPASE COLIPASE COMPLEX INHIBITED BY A C11 ALKYL PHOSPHONATE | | Descriptor: | CALCIUM ION, COLIPASE, LIPASE, ... | | Authors: | Egloff, M.-P, Van Tilbeurgh, H, Cambillau, C. | | Deposit date: | 1994-08-19 | | Release date: | 1994-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The 2.46 A resolution structure of the pancreatic lipase-colipase complex inhibited by a C11 alkyl phosphonate.
Biochemistry, 34, 1995
|
|
2A6E
 
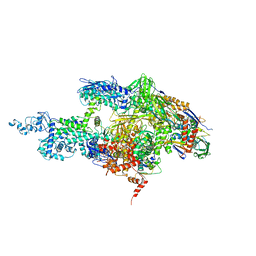 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
2A7S
 
 | | Crystal Structure of the Acyl-CoA Carboxylase, AccD5, from Mycobacterium tuberculosis | | Descriptor: | Probable propionyl-CoA carboxylase beta chain 5 | | Authors: | Lin, T, Melgar, M, Purdon, J, Tseng, T, Tsai, S.C. | | Deposit date: | 2005-07-06 | | Release date: | 2006-02-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based inhibitor design of AccD5, an essential acyl-CoA carboxylase carboxyltransferase domain of Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1LQ0
 
 | |
2ADJ
 
 | | Crystal structure of monoclonal anti-CD4 antibody Q425 in complex with Calcium | | Descriptor: | CALCIUM ION, Q425 Fab Heavy chain, Q425 Fab Light chain | | Authors: | Zhou, T, Hamer, D.H, Hendrickson, W.A, Sattentau, Q.J, Kwong, P.D. | | Deposit date: | 2005-07-20 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interfacial metal and antibody recognition.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2A8P
 
 | | 2.7 Angstrom Crystal Structure of the Complex Between the Nuclear SnoRNA Decapping Nudix Hydrolase X29 and Manganese | | Descriptor: | MANGANESE (II) ION, U8 snoRNA-binding protein X29 | | Authors: | Scarsdale, J.N, Peculis, B.A, Wright, H.T. | | Deposit date: | 2005-07-08 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of U8 snoRNA decapping nudix hydrolase, X29, and its metal and cap complexes
Structure, 14, 2006
|
|
