5JPL
 
 | | LP2006, a handcuff-topology lasso peptide antibiotic | | Descriptor: | Uncharacterized protein | | Authors: | Tietz, J.I, Schwalen, C.J, Blair, P.M, Zakai, U.I, Mitchell, D.A. | | Deposit date: | 2016-05-03 | | Release date: | 2017-03-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A new genome-mining tool redefines the lasso peptide biosynthetic landscape.
Nat. Chem. Biol., 13, 2017
|
|
5JQN
 
 | |
2LIE
 
 | | NMR structure of the lectin CCL2 | | Descriptor: | CCL2 lectin | | Authors: | Schubert, M, Walti, M.A, Egloff, P, Bleuler-Martinez, S, Aebi, M, Allain, F.F.-H, Kunzler, M. | | Deposit date: | 2011-08-29 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Plasticity of the beta-Trefoil Protein Fold in the Recognition and Control of Invertebrate Predators and Parasites by a Fungal Defence System
Plos Pathog., 8, 2012
|
|
2LIQ
 
 | | Solution structure of CCL2 in complex with glycan | | Descriptor: | CCL2 lectin, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside | | Authors: | Schubert, M, Bleuler-Martinez, S, Walti, M.A, Egloff, P, Aebi, M, Kuenzler, M, Allain, F.H.-T. | | Deposit date: | 2011-08-30 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Plasticity of the beta-Trefoil Protein Fold in the Recognition and Control of Invertebrate Predators and Parasites by a Fungal Defence System
Plos Pathog., 8, 2012
|
|
5JZW
 
 | |
6TJ3
 
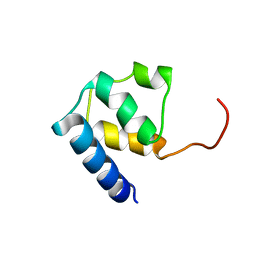 | |
5JTM
 
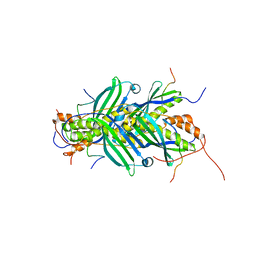 | | The structure of chaperone SecB in complex with unstructured PhoA binding site a | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5JUL
 
 | | Near atomic structure of the Dark apoptosome | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apaf-1 related killer DARK | | Authors: | Cheng, T.C, Akey, I.V, Yuan, S, Yu, Z, Ludtke, S.J, Akey, C.W. | | Deposit date: | 2016-05-10 | | Release date: | 2017-02-22 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A Near-Atomic Structure of the Dark Apoptosome Provides Insight into Assembly and Activation.
Structure, 25, 2017
|
|
5JTQ
 
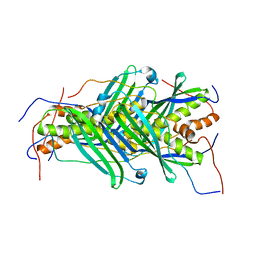 | | The structure of chaperone SecB in complex with unstructured MBP binding site d | | Descriptor: | Maltose-binding periplasmic protein, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5JZT
 
 | |
6NRB
 
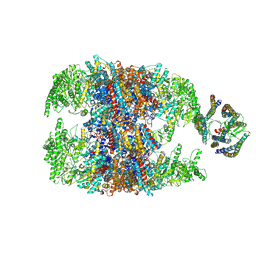 | | hTRiC-hPFD Class2 | | Descriptor: | Prefoldin subunit 1, Prefoldin subunit 2, Prefoldin subunit 3, ... | | Authors: | Gestaut, D.R, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | Deposit date: | 2019-01-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
6CPJ
 
 | | Solution structure of SH3 domain from Shank2 | | Descriptor: | SH3 and multiple ankyrin repeat domains protein 2 | | Authors: | Ishida, H, Vogel, H.J. | | Deposit date: | 2018-03-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SH3 domains from Shank scaffold proteins and their interactions with Cav1.3 calcium channels.
FEBS Lett., 592, 2018
|
|
6CPI
 
 | | Solution structure of SH3 domain from Shank1 | | Descriptor: | SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Ishida, H, Vogel, H.J. | | Deposit date: | 2018-03-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SH3 domains from Shank scaffold proteins and their interactions with Cav1.3 calcium channels.
FEBS Lett., 592, 2018
|
|
4U4F
 
 | | Structure of GluA2* in complex with partial agonist (S)-5-Nitrowillardiine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(5-nitro-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-L-alanine, ... | | Authors: | Yelshanskaya, M.V, Li, M, Sobolevsky, A.I. | | Deposit date: | 2014-07-23 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.79 Å) | | Cite: | Structure of an agonist-bound ionotropic glutamate receptor.
Science, 345, 2014
|
|
3GUB
 
 | | Crystal structure of DAPKL93G complexed with N6-(2-Phenylethyl)adenosine | | Descriptor: | 9-alpha-L-lyxofuranosyl-N-(2-phenylethyl)-9H-purin-6-amine, Death-associated protein kinase 1, SULFATE ION | | Authors: | McNamara, L.K, Schumacher, A.M, Schavocky, J.S, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2009-03-29 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structures of the DAPK gatekeeper mutant complexed with N6-modified adenosine analogs.
To be Published
|
|
1W1N
 
 | | The solution structure of the FATC Domain of the Protein Kinase TOR1 from yeast | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE TOR1 | | Authors: | Dames, S.A, Mulet, J.M, Rathgeb-Szabo, K, Hall, M.N, Grzesiek, S. | | Deposit date: | 2004-06-23 | | Release date: | 2005-03-16 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the FATC domain of the protein kinase target of rapamycin suggests a role for redox-dependent structural and cellular stability.
J. Biol. Chem., 280, 2005
|
|
3GU8
 
 | | Crystal structure of DAPKL93G with N6-cyclopentyladenosine | | Descriptor: | Death-associated protein kinase 1, N6-cyclopentyladenosine | | Authors: | McNamara, L.K, Schumacher, A.M, Schavocky, J.S, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2009-03-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of the DAPK gatekeeper mutant with N6-modified adenosine analogs.
To be Published
|
|
1I11
 
 | | SOLUTION STRUCTURE OF THE DNA BINDING DOMAIN, SOX-5 HMG BOX FROM MOUSE | | Descriptor: | TRANSCRIPTION FACTOR SOX-5 | | Authors: | Cary, P.D, Read, C.M, Davis, B, Driscoll, P.C, Crane-Robinson, C. | | Deposit date: | 2001-01-30 | | Release date: | 2001-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the DNA-binding domain of mouse Sox-5.
Protein Sci., 10, 2001
|
|
2KTV
 
 | |
2KZ1
 
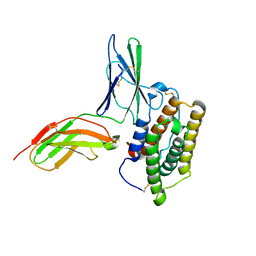 | | Inter-molecular interactions in a 44 kDa interferon-receptor complex detected by asymmetric back-protonation and 2D NOESY | | Descriptor: | Interferon alpha-2, Soluble IFN alpha/beta receptor | | Authors: | Nudelman, I, Akabayov, S.R, Schnur, E, Biron, Z, Levy, R, Xu, Y, Yang, D, Anglister, J. | | Deposit date: | 2010-06-10 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Intermolecular interactions in a 44 kDa interferon-receptor complex detected by asymmetric reverse-protonation and two-dimensional NOESY
Biochemistry, 49, 2010
|
|
4Z0I
 
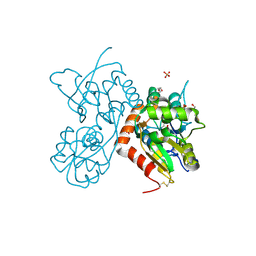 | | Crystal structure of a tetramer of GluA2 ligand binding domains bound with glutamate at 1.45 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTAMIC ACID, Glutamate receptor 2,Glutamate receptor 2, ... | | Authors: | Baranovic, J, Chebli, M, Salazar, H, Carbone, A.L, Ghisi, V, Faelber, K, Lau, A.Y, Daumke, O, Plested, A.J.R. | | Deposit date: | 2015-03-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the tetrameric wt GluA2 ligand-binding domain bound to glutamate at 1.45 Angstroms resolution
To Be Published
|
|
4FR5
 
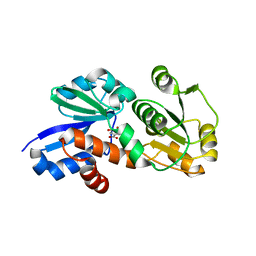 | | Crystal Structure of Shikimate Dehydrogenase (aroE) Y210S Mutant from Helicobacter pylori in Complex with Shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, Shikimate dehydrogenase | | Authors: | Cheng, W.C, Chen, T.J, Wang, W.C. | | Deposit date: | 2012-06-26 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: |
|
|
5CWG
 
 | |
4FQ8
 
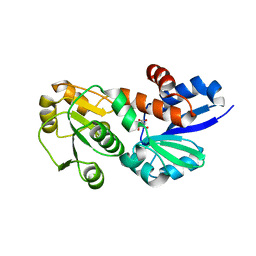 | | Crystal Structure of Shikimate Dehydrogenase (aroE) Y210A Mutant from Helicobacter pylori in Complex with Shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, Shikimate dehydrogenase | | Authors: | Cheng, W.C, Chen, T.J, Wang, W.C. | | Deposit date: | 2012-06-25 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: |
|
|
2BWM
 
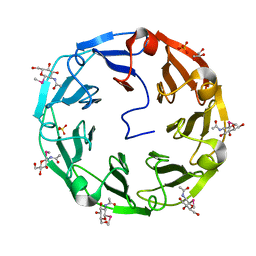 | | 1.8A CRYSTAL STRUCTURE OF of Psathyrella velutina LECTIN IN COMPLEX WITH METHYL 2-ACETAMIDO-1,2-DIDEOXY-1-SELENO-BETA-D-GLUCOPYRANOSIDE | | Descriptor: | CALCIUM ION, GLYCEROL, PSATHYRELLA VELUTINA LECTIN PVL, ... | | Authors: | Cioci, G, Mitchell, E.P, Chazalet, V, Gautier, C, Oscarson, S, Debray, H, Perez, S, Imberty, A. | | Deposit date: | 2005-07-15 | | Release date: | 2006-01-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Beta-Propeller Crystal Structure of Psathyrella Velutina Lectin: An Integrin-Like Fungal Protein Interacting with Monosaccharides and Calcium.
J.Mol.Biol., 357, 2006
|
|
