5MXB
 
 | | Crystal structure of yellow lupin LLPR-10.2B protein in complex with melatonin | | Descriptor: | Class 10 plant pathogenesis-related protein, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, SODIUM ION, ... | | Authors: | Sliwiak, J, Sikorski, M, Jaskolski, M. | | Deposit date: | 2017-01-22 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | PR-10 proteins as potential mediators of melatonin-cytokinin cross-talk in plants: crystallographic studies of LlPR-10.2B isoform from yellow lupine.
FEBS J., 285, 2018
|
|
5MXW
 
 | | Crystal structure of yellow lupin LLPR-10.2B protein in complex with melatonin and trans-zeatin. | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, Class 10 plant pathogenesis-related protein, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, ... | | Authors: | Sliwiak, J, Sikorski, M, Jaskolski, M. | | Deposit date: | 2017-01-25 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | PR-10 proteins as potential mediators of melatonin-cytokinin cross-talk in plants: crystallographic studies of LlPR-10.2B isoform from yellow lupine.
FEBS J., 285, 2018
|
|
1X7P
 
 | |
7BG7
 
 | | HRV14 in complex with its receptor ICAM-1 | | Descriptor: | Genome polyprotein, Intercellular adhesion molecule 1 | | Authors: | Hrebik, D, Fuzik, T, Plevka, P. | | Deposit date: | 2021-01-06 | | Release date: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | ICAM-1 induced rearrangements of capsid and genome prime rhinovirus 14 for activation and uncoating.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5CFC
 
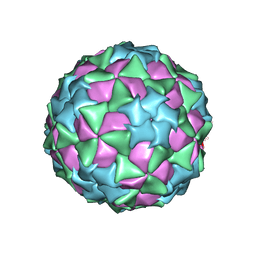 | |
3RVD
 
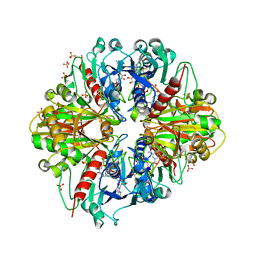 | | Crystal structure of the binary complex, obtained by soaking, of photosyntetic a4 glyceraldehyde 3-phosphate dehydrogenase (gapdh) with cp12-2, both from arabidopsis thaliana. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Thumiger, A, Falini, G, Marri, L, Sparla, F, Trost, P. | | Deposit date: | 2011-05-06 | | Release date: | 2012-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational Selection and Folding-upon-binding of Intrinsically Disordered Protein CP12 Regulate Photosynthetic Enzymes Assembly.
J.Biol.Chem., 287, 2012
|
|
2W69
 
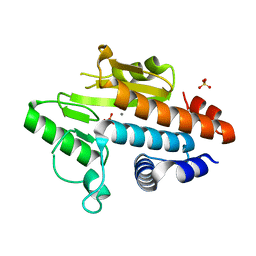 | | Influenza polymerase fragment | | Descriptor: | MANGANESE (II) ION, POLYMERASE ACIDIC PROTEIN, SULFATE ION | | Authors: | Dias, A, Bouvier, D, Crepin, T, McCarthy, A.A, Hart, D.J, Baudin, F, Cusack, S, Ruigrok, R.W.H. | | Deposit date: | 2008-12-17 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The CAP-Snatching Endonuclease of Influenza Virus Polymerase Resides in the Pa Subunit
Nature, 458, 2009
|
|
5UQJ
 
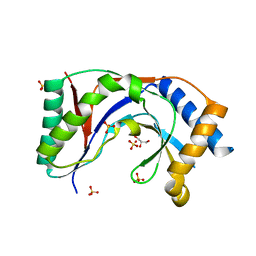 | | Structure of yeast Usb1 | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Didychuk, A.L, Montemayor, E.J, Butcher, S.E. | | Deposit date: | 2017-02-08 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Usb1 controls U6 snRNP assembly through evolutionarily divergent cyclic phosphodiesterase activities.
Nat Commun, 8, 2017
|
|
6PVK
 
 | | Bacterial 45SRbgA ribosomal particle class A | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Ortega, J, Seffouh, A, Jain, N, Jahagirdar, D, Basu, K, Razi, A, Ni, X, Guarne, A, Britton, R.A. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural consequences of the interaction of RbgA with a 50S ribosomal subunit assembly intermediate.
Nucleic Acids Res., 47, 2019
|
|
6ZV3
 
 | | TFIIS N-terminal domain (TND) from human MED26 | | Descriptor: | Mediator of RNA polymerase II transcription subunit 26 | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
6FY1
 
 | | Crystal structure of a V2p-reactive RV144 vaccine-like antibody, CAP228-16H, in complex with a scaffolded autologous V1V2 | | Descriptor: | CAP228 Autologous Scaffolded V1V2, CAP228-16H Heavy Chain, CAP228-16H Light Chain | | Authors: | Wibmer, C.K, Moore, P.L, Morris, L. | | Deposit date: | 2018-03-10 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.132 Å) | | Cite: | Common helical V1V2 conformations of HIV-1 Envelope expose the alpha 4 beta 7 binding site on intact virions.
Nat Commun, 9, 2018
|
|
6FY2
 
 | | Crystal structure of a V2p-reactive RV144 vaccine-like antibody, CAP228-16H, in complex with a heterologous CAP225 V1V2 | | Descriptor: | CAP225 Scaffolded V1V2, CAP228-16H Heavy Chain, CAP228-16H Light Chain, ... | | Authors: | Wibmer, C.K, Moore, P.L, Morris, L. | | Deposit date: | 2018-03-10 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Common helical V1V2 conformations of HIV-1 Envelope expose the alpha 4 beta 7 binding site on intact virions.
Nat Commun, 9, 2018
|
|
1KHM
 
 | | C-TERMINAL KH DOMAIN OF HNRNP K (KH3) | | Descriptor: | PROTEIN (HNRNP K) | | Authors: | Baber, J, Libutti, D, Levens, D, Tjandra, N. | | Deposit date: | 1999-01-07 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High precision solution structure of the C-terminal KH domain of heterogeneous nuclear ribonucleoprotein K, a c-myc transcription factor.
J.Mol.Biol., 289, 1999
|
|
5HDA
 
 | |
4OBC
 
 | | Crystal structure of HCV polymerase NS5b genotype 2a JFH-1 isolate with the S15G, C223H, V321I resistance mutations against the guanosine analog GS-0938 (PSI-3529238) | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Edwards, T.E, Abendroth, J, Appleby, T.C. | | Deposit date: | 2014-01-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular and Structural Basis for the Roles of Hepatitis C Virus Polymerase NS5B Amino Acids 15, 223, and 321 in Viral Replication and Drug Resistance.
Antimicrob.Agents Chemother., 58, 2014
|
|
2WFS
 
 | | Fitting of influenza virus NP structure into the 9-fold symmetryzed cryoEM reconstruction of an active RNP particle. | | Descriptor: | NUCLEOPROTEIN | | Authors: | Coloma, R, Valpuesta, J.M, Arranz, R, Carrascosa, J.L, Ortin, J, Martin-Benito, J. | | Deposit date: | 2009-04-15 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | The Structure of a Biologically Active Influenza Virus Ribonucleoprotein Complex.
Plos Pathog., 5, 2009
|
|
3NKB
 
 | | A 1.9A crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage | | Descriptor: | DNA/RNA (5'-D(*(DUR))-D(*GP*G)-R(P*CP*UP*UP*GP*CP*A)-3'), MAGNESIUM ION, The hepatitis delta virus ribozyme | | Authors: | Chen, J.-H, Yajima, R, Chadalavada, D.M, Chase, E, Bevilacqua, P.C, Golden, B.L. | | Deposit date: | 2010-06-18 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.916 Å) | | Cite: | A 1.9 A crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage.
Biochemistry, 49, 2010
|
|
1Y9X
 
 | |
2D5R
 
 | | Crystal Structure of a Tob-hCaf1 Complex | | Descriptor: | CCR4-NOT transcription complex subunit 7, Tob1 protein | | Authors: | Horiuchi, M, Suzuki, N.N, Muroya, N, Takahasi, K, Nishida, M, Yoshida, Y, Ikematsu, N, Nakamura, T, Kawamura-Tsuzuku, J, Yamamoto, T, Inagaki, F. | | Deposit date: | 2005-11-04 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the antiproliferative activity of the Tob-hCaf1 complex.
J.Biol.Chem., 284, 2009
|
|
2P7V
 
 | | Crystal structure of the Escherichia coli regulator of sigma 70, Rsd, in complex with sigma 70 domain 4 | | Descriptor: | MAGNESIUM ION, RNA polymerase sigma factor rpoD, Regulator of sigma D | | Authors: | Patikoglou, G.A, Westblade, L.F, Campbell, E.A, Lamour, V, Lane, W.J, Darst, S.A. | | Deposit date: | 2007-03-20 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the Escherichia coli regulator of sigma70, Rsd, in complex with sigma70 domain 4.
J.Mol.Biol., 372, 2007
|
|
1R08
 
 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(5-(2,6-DICHLORO-4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)PENTYL)-3-(HYDROXYETHYL OXYMETHYLENEOXYMETHYL) ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
3DCO
 
 | | Drosophila NOD (3DC4) and Bovine Tubulin (1JFF) Docked into the 11-Angstrom Cryo-EM Map of Nucleotide-Free NOD Complexed to the Microtubule | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bovine Alpha Tubulin, Bovine Beta Tubulin, ... | | Authors: | Sindelar, C.V, Cochran, J.C, Kull, F.J. | | Deposit date: | 2008-06-04 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | ATPase cycle of the nonmotile kinesin NOD allows microtubule end tracking and drives chromosome movement.
Cell(Cambridge,Mass.), 136, 2009
|
|
1MIV
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme | | Descriptor: | MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
1MIW
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
1MIY
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme in complex with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
