5WLY
 
 | | E. coli LpxH- 8 mutations | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Bohl, T.E, Aihara, H, Shi, K, Lee, J.K. | | Deposit date: | 2017-07-28 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The substrate-binding cap of the UDP-diacylglucosamine pyrophosphatase LpxH is highly flexible, enabling facile substrate binding and product release.
J. Biol. Chem., 293, 2018
|
|
4W7P
 
 | | Crystal Structure of ROCK 1 bound to YB-15-QD37 | | Descriptor: | N~1~-[2-(1H-indazol-5-yl)pyrido[3,4-d]pyrimidin-4-yl]-2-methylpropane-1,2-diamine, Rho-associated protein kinase 1 | | Authors: | Sprague, E.R. | | Deposit date: | 2014-08-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel ROCK inhibitors for the treatment of pulmonary arterial hypertension.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4W9D
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methyloxazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 3) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(4-methyl-1,3-oxazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Hewitt, S, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
4USV
 
 | |
5WVC
 
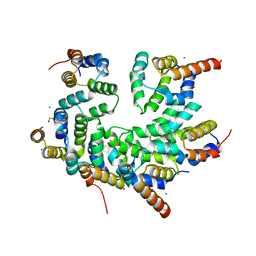 | | Structure of the CARD-CARD disk | | Descriptor: | Apoptotic protease-activating factor 1, Caspase, IODIDE ION | | Authors: | Lin, S.C, Lo, Y.C, Su, T.W. | | Deposit date: | 2016-12-24 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | Structural Insights into DD-Fold Assembly and Caspase-9 Activation by the Apaf-1 Apoptosome.
Structure, 25, 2017
|
|
1DED
 
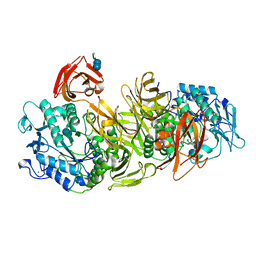 | | CRYSTAL STRUCTURE OF ALKALOPHILIC ASPARAGINE 233-REPLACED CYCLODEXTRIN GLUCANOTRANSFERASE COMPLEXED WITH AN INHIBITOR, ACARBOSE, AT 2.0 A RESOLUTION | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, CYCLODEXTRIN GLUCANOTRANSFERASE | | Authors: | Ishii, N, Haga, K, Yamane, K, Harata, K. | | Deposit date: | 1999-11-14 | | Release date: | 2000-04-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of alkalophilic asparagine 233-replaced cyclodextrin glucanotransferase complexed with an inhibitor, acarbose, at 2.0 A resolution.
J.Biochem.(Tokyo), 127, 2000
|
|
4W8I
 
 | | Crystal structure of LpSPL/Lpp2128, Legionella pneumophila sphingosine-1 phosphate lyase | | Descriptor: | Probable sphingosine-1-phosphate lyase | | Authors: | Stogios, P.J, Daniels, C, Skarina, T, Cuff, M, Di Leo, R, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-24 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Legionella pneumophila S1P-lyase targets host sphingolipid metabolism and restrains autophagy.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4W9C
 
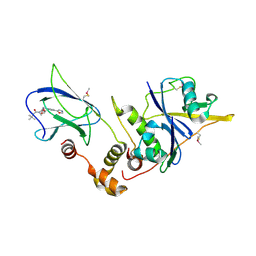 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(oxazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 2) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(1,3-oxazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Hewitt, S, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
3GNM
 
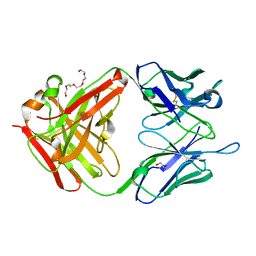 | | The crystal structure of the JAA-F11 monoclonal antibody Fab fragment | | Descriptor: | 1,2-ETHANEDIOL, JAA-F11 Fab Antibody Fragment, Heavy Chain, ... | | Authors: | Gulick, A.M, Rittenhouse-Olson, K, Jadey, S. | | Deposit date: | 2009-03-17 | | Release date: | 2010-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Computational Screening of the Human TF-Glycome Provides a Structural Definition for the Specificity of Anti-Tumor Antibody JAA-F11.
Plos One, 8, 2013
|
|
7M8Q
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit with fluorosubstituted tryptophans | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, FE (III) ION, ... | | Authors: | Johns, J.C, Banerjee, R, Shi, K, Semonis, M.M, Aihara, H, Pomerantz, W.C.K, Lipscomb, J.D. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Soluble Methane Monooxygenase Component Interactions Monitored by 19 F NMR.
Biochemistry, 60, 2021
|
|
7LP1
 
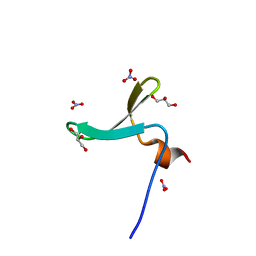 | | Structure of Nedd4L WW3 domain | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like, GLYCEROL, NITRATE ION | | Authors: | Alian, A, Alam, S.L, Thompson, T, Rheinemann, L, Sundquist, W.I. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
7LMD
 
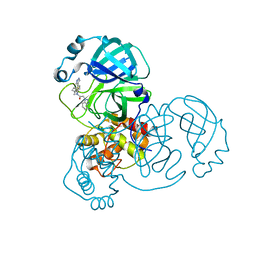 | | SARS-CoV-2 3CLPro in complex with 2-(benzotriazol-1-yl)-N-[4-(1H-pyrazol-4-yl)phenyl]-N-(3-thienylmethyl)acetamide | | Descriptor: | 2-(benzotriazol-1-yl)-~{N}-[4-(1~{H}-pyrazol-4-yl)phenyl]-~{N}-(thiophen-3-ylmethyl)ethanamide, 3C-like proteinase | | Authors: | Goins, C.M, Arya, T, Macdonald, J.D, Stauffer, S.R. | | Deposit date: | 2021-02-05 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-Based Optimization of ML300-Derived, Noncovalent Inhibitors Targeting the Severe Acute Respiratory Syndrome Coronavirus 3CL Protease (SARS-CoV-2 3CL pro ).
J.Med.Chem., 65, 2022
|
|
7M6L
 
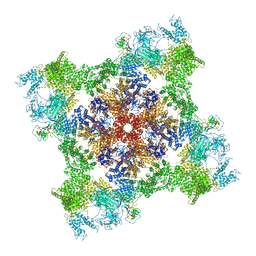 | | High resolution structure of the membrane embedded skeletal muscle ryanodine receptor | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Melville, Z, Kim, K, Clarke, O.B, Marks, A.R. | | Deposit date: | 2021-03-25 | | Release date: | 2021-08-18 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | High-resolution structure of the membrane-embedded skeletal muscle ryanodine receptor.
Structure, 30, 2022
|
|
7LMH
 
 | | SARS-CoV-1 3CLPro in complex with 2-(1H-benzo[d][1,2,3]triazol-1-yl)-N-(4-(pyridin-3-yl)phenyl)-N-(thiophen-3-ylmethyl)acetamide | | Descriptor: | 2-(benzotriazol-1-yl)-~{N}-(4-pyridin-3-ylphenyl)-~{N}-(thiophen-3-ylmethyl)ethanamide, 3C-like proteinase, GLYCEROL | | Authors: | Arya, T, Goins, C.M, Macdonald, J.D, Stauffer, S.R. | | Deposit date: | 2021-02-05 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Optimization of ML300-Derived, Noncovalent Inhibitors Targeting the Severe Acute Respiratory Syndrome Coronavirus 3CL Protease (SARS-CoV-2 3CL pro ).
J.Med.Chem., 65, 2022
|
|
7LMF
 
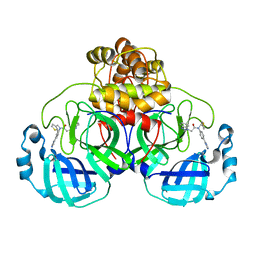 | | SARS-CoV-2 3CLPro in complex with 2-(benzotriazol-1-yl)-N-[4-(1H-imidazol-4-yl)phenyl]-N-(3-thienylmethyl)acetamide | | Descriptor: | 2-(benzotriazol-1-yl)-~{N}-[4-(1~{H}-imidazol-4-yl)phenyl]-~{N}-(thiophen-3-ylmethyl)ethanamide, 3C-like proteinase | | Authors: | Goins, C.M, Arya, T, Macdonald, J.D, Stauffer, S.R. | | Deposit date: | 2021-02-05 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Optimization of ML300-Derived, Noncovalent Inhibitors Targeting the Severe Acute Respiratory Syndrome Coronavirus 3CL Protease (SARS-CoV-2 3CL pro ).
J.Med.Chem., 65, 2022
|
|
7LPQ
 
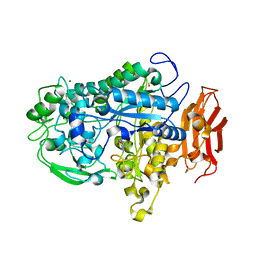 | | Crystal structure of Cryptococcus neoformans sterylglucosidase 1 with hit 9 | | Descriptor: | Cytoplasmic protein, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Pereira de Sa, N, Del Poeta, M, Airola, M.V. | | Deposit date: | 2021-02-12 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and inhibition of Cryptococcus neoformans sterylglucosidase to develop antifungal agents.
Nat Commun, 12, 2021
|
|
7LPO
 
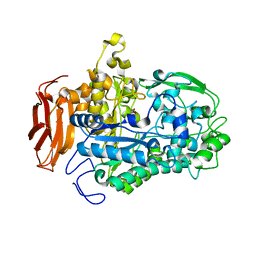 | | Crystal structure of Cryptococcus neoformans sterylglucosidase 1 with tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytoplasmic protein, MAGNESIUM ION | | Authors: | Pereira de Sa, N, Del Poeta, M, Airola, M.V. | | Deposit date: | 2021-02-12 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure and inhibition of Cryptococcus neoformans sterylglucosidase to develop antifungal agents.
Nat Commun, 12, 2021
|
|
5WUN
 
 | |
4W1V
 
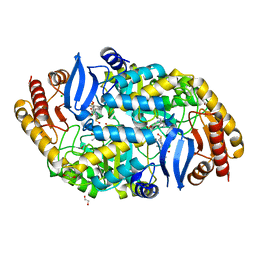 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis, complexed with a thiazole inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, CHLORIDE ION, ... | | Authors: | Finzel, B.C, Dai, R. | | Deposit date: | 2014-08-13 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Target-Based Identification of Whole-Cell Active Inhibitors of Biotin Biosynthesis in Mycobacterium tuberculosis.
Chem.Biol., 22, 2015
|
|
7M18
 
 | | HeLa-tubulin in complex with cryptophycin 1 | | Descriptor: | Cryptophycin 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Eren, E. | | Deposit date: | 2021-03-12 | | Release date: | 2021-09-08 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Conformational changes in tubulin upon binding cryptophycin-52 reveal its mechanism of action.
J.Biol.Chem., 297, 2021
|
|
7M20
 
 | | 18-mer HeLa-tubulin rings in complex with Cryptophycin 1 | | Descriptor: | Cryptophycin 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Eren, E. | | Deposit date: | 2021-03-15 | | Release date: | 2021-09-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Conformational changes in tubulin upon binding cryptophycin-52 reveal its mechanism of action.
J.Biol.Chem., 297, 2021
|
|
4W9E
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(thiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 4) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
7M6A
 
 | | High resolution structure of the membrane embedded skeletal muscle ryanodine receptor | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Melville, Z, Kim, K, Clarke, O.B, Marks, A.R. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-08 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | High-resolution structure of the membrane-embedded skeletal muscle ryanodine receptor.
Structure, 30, 2022
|
|
8CXF
 
 | |
5MVS
 
 | | Crystal structure of Dot1L in complex with adenosine and inhibitor CPD1 [N6-(2,6-dichlorophenyl)-N6-(pent-2-yn-1-yl)quinoline-4,6-diamine] | | Descriptor: | ADENOSINE, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-17 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
