1SLT
 
 | |
1WTG
 
 | | Human Factor Viia-Tissue Factor Complexed with ethylsulfonamide-D-biphenylalanine-Gln-p-aminobenzamidine | | Descriptor: | 2-(3-BIPHENYL-4-YL-2-ETHANESULFONYLAMINO-PROPIONYLAMINO)-PENTANEDIOIC ACID 5-AMIDE 1-(4-CARBAMIMIDOYL-BENZYLAMIDE), CALCIUM ION, Coagulation factor VII, ... | | Authors: | Kadono, S, Sakamoto, S, Kikuchi, Y, Oh-Eda, M, Yabuta, N, Kitazawa, K, Yoshihashi, T, Suzuki, T, Koga, T, Hattori, K, Shiraishi, T, Kodama, M, Haramura, H, Ono, Y, Esaki, T, Sato, H, Watanabe, Y, Itoh, S, Ohta, M, Kozono, T. | | Deposit date: | 2004-11-23 | | Release date: | 2005-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel interactions of large P3 moiety and small P4 moiety in the binding of the peptide mimetic factor VIIa inhibitor
Biochem.Biophys.Res.Commun., 326, 2005
|
|
1WU4
 
 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase | | Descriptor: | GLYCEROL, NICKEL (II) ION, xylanase Y | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Basis for the Specificity of the Reducing End Xylose-releasing Exo-oligoxylanase from Bacillus halodurans C-125
J.Biol.Chem., 280, 2005
|
|
2D81
 
 | | PHB depolymerase (S39A) complexed with R3HB trimer | | Descriptor: | (1R)-3-{[(1R)-3-METHOXY-1-METHYL-3-OXOPROPYL]OXY}-1-METHYL-3-OXOPROPYL (3R)-3-HYDROXYBUTANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHB depolymerase | | Authors: | Hisano, T, Kasuya, K, Saito, T, Iwata, T, Miki, K. | | Deposit date: | 2005-11-30 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The Crystal Structure of Polyhydroxybutyrate Depolymerase from Penicillium funiculosum Provides Insights into the Recognition and Degradation of Biopolyesters
J.Mol.Biol., 356, 2006
|
|
1SN1
 
 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M1 | | Descriptor: | PROTEIN (NEUROTOXIN BMK M1) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
1WWJ
 
 | | crystal structure of KaiB from Synechocystis sp. | | Descriptor: | Circadian clock protein kaiB, D-MALATE, IMIDAZOLE, ... | | Authors: | Hitomi, K, Oyama, T, Han, S, Arvai, A.S, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tetrameric architecture of the circadian clock protein KaiB. A novel interface for intermolecular interactions and its impact on the circadian rhythm.
J.Biol.Chem., 280, 2005
|
|
1SOX
 
 | | SULFITE OXIDASE FROM CHICKEN LIVER | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, MOLYBDENUM ATOM, ... | | Authors: | Kisker, C, Schindelin, H, Rees, D.C. | | Deposit date: | 1997-12-31 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of sulfite oxidase deficiency from the structure of sulfite oxidase.
Cell(Cambridge,Mass.), 91, 1997
|
|
1SQ0
 
 | | Crystal Structure of the Complex of the Wild-type Von Willebrand Factor A1 domain and Glycoprotein Ib alpha at 2.6 Angstrom Resolution | | Descriptor: | Platelet glycoprotein Ib alpha chain (Glycoprotein Ibalpha) (GP-Ib alpha) (GPIbA) (GPIb-alpha) (CD42B-alpha) (CD42B) [Contains: Glycocalicin], Von Willebrand factor (vWF) [Contains: Von Willebrand antigen II] | | Authors: | Dumas, J.J, Kumar, R, McDonagh, T, Sullivan, F, Stahl, M.L, Somers, W.S, Mosyak, L. | | Deposit date: | 2004-03-17 | | Release date: | 2004-04-13 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the wild-type von Willebrand factor A1-glycoprotein Ibalpha complex reveals conformation differences with a complex bearing von Willebrand disease mutations
J.Biol.Chem., 279, 2004
|
|
1WXW
 
 | | Crystal structure of Tt1595, a putative SAM-dependent methyltransferase from Thermus thermophillus HB8 | | Descriptor: | HEXANE-1,6-DIOL, hypothetical protein TTHA1280 | | Authors: | Pioszak, A.A, Murayama, K, Nakagawa, N, Ebihara, A, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-02 | | Release date: | 2005-08-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of a putative RNA 5-methyluridine methyltransferase, Thermus thermophilus TTHA1280, and its complex with S-adenosyl-L-homocysteine.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1WWI
 
 | | Crystal structure of ttk003001566 from Thermus Thermophilus HB8 | | Descriptor: | hypothetical protein TTHA1479 | | Authors: | Wang, H, Murayama, K, Terada, T, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-05 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of ttk003001566 from Thermus Thermophilus HB8
TO BE PUBLISHED
|
|
1WWR
 
 | | Crystal structure of tRNA adenosine deaminase TadA from Aquifex aeolicus | | Descriptor: | ZINC ION, tRNA adenosine deaminase TadA | | Authors: | Kuratani, M, Ishii, R, Bessho, Y, Fukunaga, R, Sengoku, T, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-12 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of tRNA Adenosine Deaminase (TadA) from Aquifex aeolicus
J.Biol.Chem., 280, 2005
|
|
1SOM
 
 | | TORPEDO CALIFORNICA ACETYLCHOLINESTERASE INHIBITED BY NERVE AGENT GD (SOMAN). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, METHYLPHOSPHONIC ACID ESTER GROUP, PROTEIN (ACETYLCHOLINESTERASE), ... | | Authors: | Greenblatt, H.M, Millard, C.B, Sussman, J.L, Silman, I. | | Deposit date: | 1999-03-17 | | Release date: | 1999-06-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of aged phosphonylated acetylcholinesterase: nerve agent reaction products at the atomic level.
Biochemistry, 38, 1999
|
|
1QWA
 
 | | NMR structure of 5'-r(GGAUGCCUCCCGAGUGCAUCC): an RNA hairpin derived from the mouse 5'ETS that binds nucleolin RBD12. | | Descriptor: | 18S ribosomal RNA, 5'ETS | | Authors: | Finger, L.D, Trantirek, L, Johansson, C, Feigon, J. | | Deposit date: | 2003-09-01 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Strucutres of Stem-loop RNAs that Bind to the Two N-terminal RNA Binding Domains of Nucleolin
Nucleic Acids Res., 31, 2003
|
|
2D2M
 
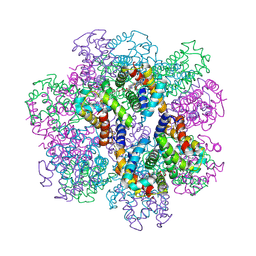 | | Structure of an extracellular giant hemoglobin of the gutless beard worm Oligobrachia mashikoi | | Descriptor: | Giant hemoglobin, A1(b) globin chain, A2(a5) globin chain, ... | | Authors: | Numoto, N, Nakagawa, T, Kita, A, Sasayama, Y, Fukumori, Y, Miki, K. | | Deposit date: | 2005-09-12 | | Release date: | 2005-10-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of an extracellular giant hemoglobin of the gutless beard worm Oligobrachia mashikoi.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1SPD
 
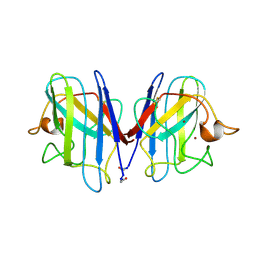 | |
1WZ2
 
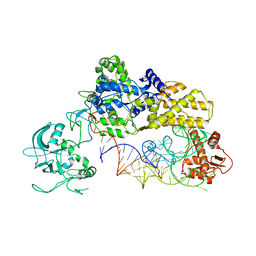 | |
2D3Z
 
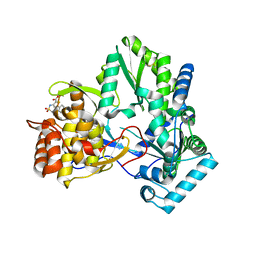 | | X-ray crystal structure of hepatitis C virus RNA-dependent RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | 5-(4-FLUOROPHENYL)-3-{[(4-METHYLPHENYL)SULFONYL]AMINO}THIOPHENE-2-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-04 | | Release date: | 2006-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
1SQH
 
 | | X-RAY STRUCTURE OF DROSOPHILA MALONOGASTER PROTEIN Q9VR51 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET FR87. | | Descriptor: | hypothetical protein CG14615-PA | | Authors: | Kuzin, A.P, Chen, Y, Forouhar, F, Acton, T.B, Xiao, R, Cooper, B, Ho, C.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-18 | | Release date: | 2004-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-RAY STRUCTURE OF DROSOPHILA MALONOGASTER PROTEIN Q9VR51 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET FR87
To be Published
|
|
1X0M
 
 | | a Human Kynurenine Aminotransferase II Homologue from Pyrococcus horikoshii OT3 | | Descriptor: | Aminotransferase II Homologue | | Authors: | Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2005-03-24 | | Release date: | 2005-04-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a human kynurenine aminotransferase II homologue from Pyrococcus horikoshii OT3 at 2.20 A resolution
Proteins, 61, 2005
|
|
2D5R
 
 | | Crystal Structure of a Tob-hCaf1 Complex | | Descriptor: | CCR4-NOT transcription complex subunit 7, Tob1 protein | | Authors: | Horiuchi, M, Suzuki, N.N, Muroya, N, Takahasi, K, Nishida, M, Yoshida, Y, Ikematsu, N, Nakamura, T, Kawamura-Tsuzuku, J, Yamamoto, T, Inagaki, F. | | Deposit date: | 2005-11-04 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the antiproliferative activity of the Tob-hCaf1 complex.
J.Biol.Chem., 284, 2009
|
|
1SBH
 
 | | SUBTILISIN BPN' 8397+1 (E.C. 3.4.21.14) MUTANT (M50F, N76D, G169A, Q206C, N218S, K256Y) | | Descriptor: | CALCIUM ION, SUBTILISIN 8397+1 | | Authors: | Kidd, R.D, Farber, G.K. | | Deposit date: | 1995-09-01 | | Release date: | 1995-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A weak calcium binding site in subtilisin BPN' has a dramatic effect on protein stability.
J.Am.Chem.Soc., 118, 1996
|
|
1WC4
 
 | | Soluble adenylyl cyclase CyaC from S. platensis in complex with alpha, beta-methylene-ATP and Europium | | Descriptor: | ADENYLATE CYCLASE, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, EUROPIUM (III) ION | | Authors: | Steegborn, C, Litvin, T.N, Levin, L.R, Buck, J, Wu, H. | | Deposit date: | 2004-11-08 | | Release date: | 2004-12-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bicarbonate Activation of Adenylyl Cyclase Via Promotion of Catalytic Active Site Closure and Metal Recruitment
Nat.Struct.Mol.Biol., 12, 2005
|
|
1X1B
 
 | | Crystal structure of BchU complexed with S-adenosyl-L-homocysteine | | Descriptor: | CrtF-related protein, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Yamaguchi, H, Wada, K, Fukuyama, K. | | Deposit date: | 2005-04-03 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of BchU, a Methyltransferase Involved in Bacteriochlorophyll c Biosynthesis, and its Complex with S-adenosylhomocysteine: Implications for Reaction Mechanism.
J.Mol.Biol., 360, 2006
|
|
2D63
 
 | | Aspartate Aminotransferase Mutant MA With Isovaleric Acid | | Descriptor: | Aspartate aminotransferase, ISOVALERIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-09 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2D1O
 
 | | Stromelysin-1 (MMP-3) complexed to a hydroxamic acid inhibitor | | Descriptor: | CALCIUM ION, SM-25453, Stromelysin-1, ... | | Authors: | Kohno, T, Hochigai, H, Yamashita, E, Tsukihara, T, Kanaoka, M. | | Deposit date: | 2005-08-30 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structures of the catalytic domain of human stromelysin-1 (MMP-3) and collagenase-3 (MMP-13) with a hydroxamic acid inhibitor SM-25453
Biochem.Biophys.Res.Commun., 344, 2006
|
|
