1A4U
 
 | |
8AEQ
 
 | |
8AET
 
 | |
8AEO
 
 | |
8AER
 
 | |
8A7S
 
 | |
8A8T
 
 | |
8A30
 
 | |
3KM0
 
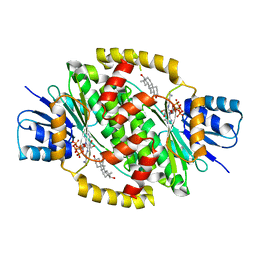 | | 17betaHSD1 in complex with 3beta-diol | | Descriptor: | 5-ALPHA-ANDROSTANE-3-BETA,17BETA-DIOL, Estradiol 17-beta-dehydrogenase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mazumdar, M, Lin, S.-X. | | Deposit date: | 2009-11-09 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of sex-steroid translation
To be Published
|
|
3KVO
 
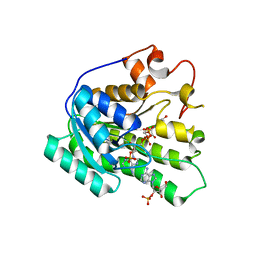 | | Crystal structure of the catalytic domain of human Hydroxysteroid dehydrogenase like 2 (HSDL2) | | Descriptor: | Hydroxysteroid dehydrogenase-like protein 2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ugochukwu, E, Bhatia, C, Huang, J, Pilka, E, Muniz, J.R.C, Pike, A.C.W, Krojer, T, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Verdin, E.M, Oppermann, U, Kavanagh, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-30 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the catalytic domain of human Hydroxysteroid dehydrogenase like 2 (HSDL2)
To be Published
|
|
3KZV
 
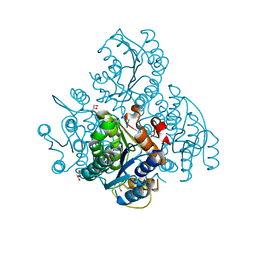 | | The crystal structure of a cytoplasmic protein with unknown function from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Uncharacterized oxidoreductase YIR035C | | Authors: | Zhang, R, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a cytoplasmic protein with unknown function from Saccharomyces cerevisiae
To be Published
|
|
3L77
 
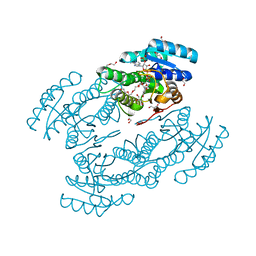 | | X-ray structure alcohol dehydrogenase from archaeon Thermococcus sibiricus complexed with 5-hydroxy-NADP | | Descriptor: | 1,2-ETHANEDIOL, 5-hydroxy-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, GLYCEROL, ... | | Authors: | Lyashenko, A.V, Lashkov, A.A, Gabdoulkhakov, A.G, Mikhailov, A.M. | | Deposit date: | 2009-12-28 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray structure alcohol dehydrogenase from archaeon Thermococcus sibiricus complexed with 5-hydroxy-NADP
To be Published
|
|
3LF1
 
 | |
3L6E
 
 | | Crystal structure of putative short chain dehydrogenase/reductase family oxidoreductase from Aeromonas hydrophila subsp. hydrophila ATCC 7966 | | Descriptor: | Oxidoreductase, short-chain dehydrogenase/reductase family, SULFATE ION | | Authors: | Malashkevich, V.N, Patskovsky, Y, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-23 | | Release date: | 2010-02-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of putative short chain dehydrogenase/reductase family oxidoreductase from Aeromonas
hydrophila subsp. hydrophila ATCC 7966
To be Published
|
|
3LF2
 
 | | NADPH Bound Structure of the Short Chain Oxidoreductase Q9HYA2 from Pseudomonas aeruginosa PAO1 Containing an Atypical Catalytic Center | | Descriptor: | CHLORIDE ION, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Huether, R, Umland, T.C, Duax, W.L. | | Deposit date: | 2010-01-15 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The short-chain oxidoreductase Q9HYA2 from Pseudomonas aeruginosa PAO1 contains an atypical catalytic center.
Protein Sci., 19, 2010
|
|
3LZ6
 
 | | Guinea Pig 11beta hydroxysteroid dehydrogenase with PF-877423 | | Descriptor: | Corticosteroid 11-beta-dehydrogenase isozyme 1, N-adamantan-2-yl-1-ethyl-D-prolinamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Pauly, T.A. | | Deposit date: | 2010-03-01 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The development and SAR of pyrrolidine carboxamide 11beta-HSD1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3M1A
 
 | | The Crystal Structure of a Short-chain Dehydrogenase from Streptomyces avermitilis to 2A | | Descriptor: | ACETATE ION, Putative dehydrogenase, SODIUM ION | | Authors: | Stein, A.J, Evdokimova, E, Egorova, O, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-04 | | Release date: | 2010-03-23 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of a Short-chain Dehydrogenase from Streptomyces avermitilis to 2A
To be Published
|
|
3KLM
 
 | | 17beta-HSD1 in complex with DHT | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Estradiol 17-beta-dehydrogenase 1, GLYCEROL | | Authors: | Mazumdar, M. | | Deposit date: | 2009-11-08 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 17beta-hydroxysteroid dehydrogenase type 1 stimulates breast cancer by dihydrotestosterone inactivation in addition to estradiol production.
Mol.Endocrinol., 24, 2010
|
|
3KLP
 
 | | 17beta-HSD1 in complex with A-diol | | Descriptor: | (3alpha,8alpha,17beta)-androst-5-ene-3,17-diol, Estradiol 17-beta-dehydrogenase 1 | | Authors: | Mazumdar, M, Lin, S.-X. | | Deposit date: | 2009-11-08 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of sex-steroids: their translational activity
To be Published
|
|
6K8V
 
 | |
6L1H
 
 | |
6L1G
 
 | |
6I79
 
 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 4 | | Descriptor: | 6-[(4-~{tert}-butyl-1,3-thiazol-2-yl)methyl]-4,6-diazaspiro[2.4]heptane-5,7-dione, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Sepiapterin reductase | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-16 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6I6T
 
 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 5 | | Descriptor: | 1,2-ETHANEDIOL, 4-bromanyl-1-oxidanyl-naphthalene-2-carboxylic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6K8U
 
 | |
