1QKT
 
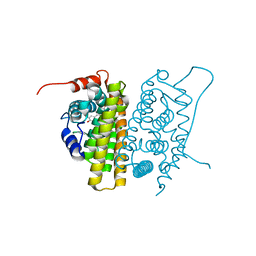 | | MUTANT ESTROGEN NUCLEAR RECEPTOR LIGAND BINDING DOMAIN COMPLEXED WITH ESTRADIOL | | Descriptor: | ESTRADIOL, ESTRADIOL RECEPTOR | | Authors: | Ruff, M, Gangloff, M, Eiler, S, Duclaud, S, Wurtz, J.M, Moras, D. | | Deposit date: | 1999-08-05 | | Release date: | 2000-08-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Mutant Heralpha Ligand- Binding Domain Reveals Key Structural Features for the Mechanism of Partial Agonism
J.Biol.Chem., 276, 2001
|
|
4UTW
 
 | | Structural characterisation of NanE, ManNac6P C2 epimerase, from Clostridium perfingens | | Descriptor: | CHLORIDE ION, N-acetyl-D-glucosamine-6-phosphate, PUTATIVE N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE | | Authors: | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway
J.Biol.Chem., 289, 2014
|
|
4F27
 
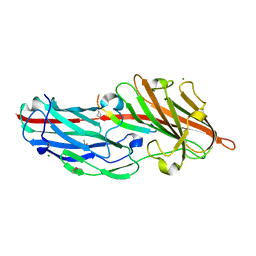 | | Crystal structures reveal the multi-ligand binding mechanism of the Staphylococcus aureus ClfB | | Descriptor: | Clumping factor B, MAGNESIUM ION, peptide from Fibrinogen alpha chain | | Authors: | Yang, M.J, Xiang, H, Wang, J.W, Liu, B, Chen, Y.G, Liu, L, Deng, X.M, Feng, Y. | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.917 Å) | | Cite: | Crystal Structures Reveal the Multi-Ligand Binding Mechanism of Staphylococcus aureus ClfB
Plos Pathog., 8, 2012
|
|
184L
 
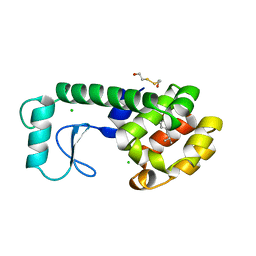 | |
1P8D
 
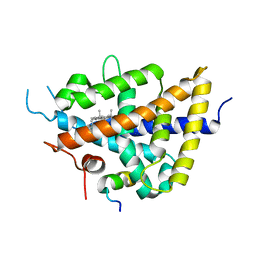 | | X-Ray Crystal Structure of LXR Ligand Binding Domain with 24(S),25-epoxycholesterol | | Descriptor: | 17-[3-(3,3-DIMETHYL-OXIRANYL)-1-METHYL-PROPYL]-10,13-DIMETHYL-2,3,4,7,8,9,10,11,12,13,14,15,16,17-TETRADECAHYDRO-1H-CYC LOPENTA[A]PHENANTHREN-3-OL, Oxysterols receptor LXR-beta, nuclear receptor coactivator 1 isoform 3 | | Authors: | Williams, S, Bledsoe, R.K, Collins, J.L, Boggs, S, Lambert, M.H, Miller, A.B, Moore, J, McKee, D.D, Moore, L, Nichols, J, Parks, D, Watson, M, Wisely, B, Willson, T.M. | | Deposit date: | 2003-05-06 | | Release date: | 2003-07-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystal structure of the liver X receptor beta ligand binding domain: regulation by
a histidine-tryptophan switch.
J.Biol.Chem., 278, 2003
|
|
6U7P
 
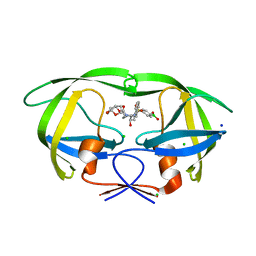 | | HIV-1 wild type protease with GRL-03119A, with phenyl-boronic-acid as P2'-ligand and with a hexahydro-4H-furo-pyran as the P2-ligand | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Kneller, D.W, Weber, I.T. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Potent HIV-1 Protease Inhibitors Containing Carboxylic and Boronic Acids: Effect on Enzyme Inhibition and Antiviral Activity and Protein-Ligand X-ray Structural Studies.
Chemmedchem, 14, 2019
|
|
6U7O
 
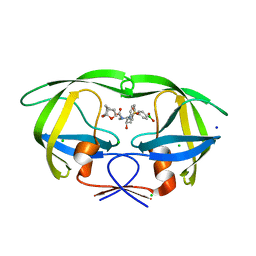 | | HIV-1 wild type protease with GRL-00819A, with phenyl-boronic-acid as P2'-ligand and with a 6-5-5-ring fused crown-like tetrahydropyranofuran as the P2-ligand | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Kneller, D.W, Weber, I.T. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Potent HIV-1 Protease Inhibitors Containing Carboxylic and Boronic Acids: Effect on Enzyme Inhibition and Antiviral Activity and Protein-Ligand X-ray Structural Studies.
Chemmedchem, 14, 2019
|
|
4F20
 
 | | Crystal structures reveal the multi-ligand binding mechanism of the Staphylococcus aureus ClfB | | Descriptor: | Clumping factor B, MAGNESIUM ION, peptide from Dermokine | | Authors: | Yang, M.J, Xiang, H, Wang, J.W, Liu, B, Chen, Y.G, Liu, L, Deng, X.M, Feng, Y. | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal Structures Reveal the Multi-Ligand Binding Mechanism of Staphylococcus aureus ClfB
Plos Pathog., 8, 2012
|
|
2LUI
 
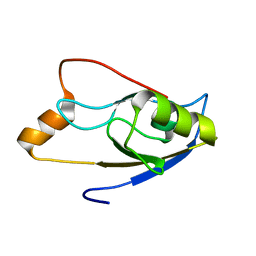 | | Structure of the PICK PDZ domain in complex with the DAT C-terminal | | Descriptor: | PICK1 PDZ DOMAIN FUSED TO THE C10 DAT LIGAND | | Authors: | Erlendsson, S, Rathje, M, Heidarsson, P.O, Poulsen, F.M, Madsen, K.L, Teilum, K, Gether, U. | | Deposit date: | 2012-06-14 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Protein interacting with C-kinase 1 (PICK1) binding promiscuity relies on unconventional PSD-95/discs-large/ZO-1 homology (PDZ) binding modes for nonclass II PDZ ligands.
J.Biol.Chem., 289, 2014
|
|
185L
 
 | |
4WVS
 
 | | Crystal structure of XIAP-BIR2 domain complexed with (S)-3-(4-methoxyphenyl)-2-((S)-2-((S)-1-((S)-2-((S)-2-(methylamino)propanamido)pent-4-ynoyl)pyrrolidine-2-carboxamido)-3-phenylpropanamido)propanoic acid | | Descriptor: | 3,11-DIFLUORO-6,8,13-TRIMETHYL-8H-QUINO[4,3,2-KL]ACRIDIN-13-IUM, E3 ubiquitin-protein ligase XIAP, GLYCEROL, ... | | Authors: | Pokross, M.E. | | Deposit date: | 2014-11-07 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The Discovery of Macrocyclic XIAP Antagonists from a DNA-Programmed Chemistry Library, and Their Optimization To Give Lead Compounds with in Vivo Antitumor Activity.
J.Med.Chem., 58, 2015
|
|
1LAF
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | ARGININE, LYSINE, ORNITHINE-BINDING PROTEIN | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
1LAG
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | HISTIDINE, LYSINE, ARGININE, ... | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
1LAH
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | L-ornithine, LYSINE, ARGININE, ... | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
6HWP
 
 | |
8GCB
 
 | | Structure of RNF125 in complex with a UbcH5b~Ub conjugate | | Descriptor: | E3 ubiquitin-protein ligase RNF125, Ubiquitin-conjugating enzyme E2 D2, ZINC ION | | Authors: | Middleton, A.J, Day, C.L, Fokkens, T.J. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Zinc finger 1 of the RING E3 ligase, RNF125, interacts with the E2 to enhance ubiquitylation.
Structure, 31, 2023
|
|
181L
 
 | |
188L
 
 | |
8GBQ
 
 | | Structure of RNF125 in complex with UbcH5b | | Descriptor: | E3 ubiquitin-protein ligase RNF125, GLYCEROL, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Middleton, A.J, Day, C.L, Fokkens, T.J. | | Deposit date: | 2023-02-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Zinc finger 1 of the RING E3 ligase, RNF125, interacts with the E2 to enhance ubiquitylation.
Structure, 31, 2023
|
|
186L
 
 | |
1IKJ
 
 | | 1.27 A CRYSTAL STRUCTURE OF NITROPHORIN 4 FROM RHODNIUS PROLIXUS COMPLEXED WITH IMIDAZOLE | | Descriptor: | IMIDAZOLE, NITROPHORIN 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roberts, S.A, Weichsel, A, Qui, Y, Shelnutt, J.A, Walker, F.A, Montfort, W.R. | | Deposit date: | 2001-05-03 | | Release date: | 2001-10-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Ligand-induced heme ruffling and bent no geometry in ultra-high-resolution structures of nitrophorin 4.
Biochemistry, 40, 2001
|
|
187L
 
 | |
5UPQ
 
 | | Acyl-CoA synthetase PtmA2 from Streptomyces platensis in complex with SBNP465 ligand | | Descriptor: | 5'-O-[(R)-{[(7beta,8alpha,9beta,10alpha,13alpha,16beta)-7,16-dihydroxy-18-oxokauran-18-yl]oxy}(hydroxy)phosphoryl]adenosine, Acyl-CoA synthetase PtmA2, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J.D, Chang, C.Y, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-02-03 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Natural separation of the acyl-CoA ligase reaction results in a non-adenylating enzyme.
Nat. Chem. Biol., 14, 2018
|
|
1BTJ
 
 | | HUMAN SERUM TRANSFERRIN, RECOMBINANT N-TERMINAL LOBE, APO FORM, CRYSTAL FORM 2 | | Descriptor: | PROTEIN (SERUM TRANSFERRIN) | | Authors: | Jeffrey, P.D, Bewley, M.C, Macgillivray, R.T.A, Mason, A.B, Woodworth, R.C, Baker, E.N. | | Deposit date: | 1998-09-01 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Ligand-induced conformational change in transferrins: crystal structure of the open form of the N-terminal half-molecule of human transferrin.
Biochemistry, 37, 1998
|
|
182L
 
 | |
