1Z70
 
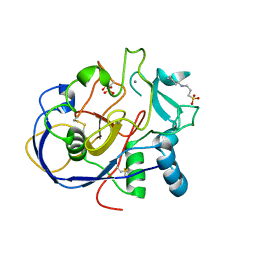 | | 1.15A resolution structure of the formylglycine generating enzyme FGE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, C-alpha-formyglycine-generating enzyme, ... | | Authors: | Rudolph, M.G. | | Deposit date: | 2005-03-23 | | Release date: | 2005-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | De novo calcium/sulfur SAD phasing of the human formylglycine-generating enzyme using in-house data.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1SXC
 
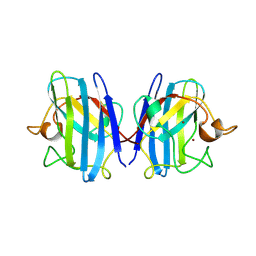 | | CRYSTAL STRUCTURE OF REDUCED BOVINE ERYTHROCYTE SUPEROXIDE DISMUTASE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Rypniewski, W.R, Mangani, S, Bruni, B, Orioli, P, Casati, M, Wilson, K.S. | | Deposit date: | 1995-03-17 | | Release date: | 1995-06-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of reduced bovine erythrocyte superoxide dismutase at 1.9 A resolution.
J.Mol.Biol., 251, 1995
|
|
3OMN
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with D132A mutation in the reduced state | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OM3
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with K362M mutation in the reduced state | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-26 | | Release date: | 2011-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4LQ1
 
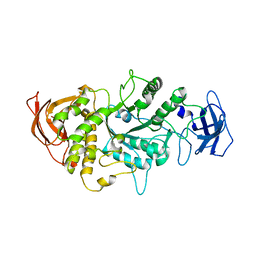 | |
4H96
 
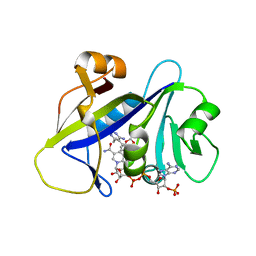 | | Candida albicans dihydrofolate reductase complexed with NADPH and 5-{3-[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-methoxyphenyl]prop-1-yn-1-yl}-6-ethylpyrimidine-2,4-diamine (UCP1018) | | Descriptor: | 5-{3-[3-(1,3-benzodioxol-5-yl)-5-methoxyphenyl]prop-1-yn-1-yl}-6-ethylpyrimidine-2,4-diamine, Dihydrofolate Reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Paulsen, J.L, Anderson, A.C. | | Deposit date: | 2012-09-24 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of the active sites of dihydrofolate reductase from two species of Candida uncovers ligand-induced conformational changes shared among species.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1A0M
 
 | | 1.1 ANGSTROM CRYSTAL STRUCTURE OF A-CONOTOXIN [TYR15]-EPI | | Descriptor: | ALPHA-CONOTOXIN [TYR15]-EPI | | Authors: | Hu, S.-H, Loughnan, M, Miller, R, Weeks, C.M, Blessing, R.H, Alewood, P.F, Lewis, R.J, Martin, J.L. | | Deposit date: | 1997-12-03 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1 A resolution crystal structure of [Tyr15]EpI, a novel alpha-conotoxin from Conus episcopatus, solved by direct methods.
Biochemistry, 37, 1998
|
|
2BR0
 
 | | DNA Adduct Bypass Polymerization by Sulfolobus solfataricus Dpo4. Analysis and Crystal Structures of Multiple Base-Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*C)-3', 5'-D(*TP*CP*AP*CP*GNEP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2005-04-28 | | Release date: | 2005-06-23 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | DNA Adduct Bypass Polymerization by Sulfolobus Solfataricus DNA Polymerase Dpo4: Analysis and Crystal Structures of Multiple Base Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine.
J.Biol.Chem., 280, 2005
|
|
3V9I
 
 | |
2RDT
 
 | | Crystal Structure of Human Glycolate Oxidase (GO) in Complex with CDST | | Descriptor: | 5-(dodecylthio)-1H-1,2,3-triazole-4-carboxylic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Murray, M.S, Holmes, R.P, Lowther, W.T. | | Deposit date: | 2007-09-25 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Active Site and Loop 4 Movements within Human Glycolate Oxidase: Implications for Substrate Specificity and Drug Design.
Biochemistry, 47, 2008
|
|
2ANQ
 
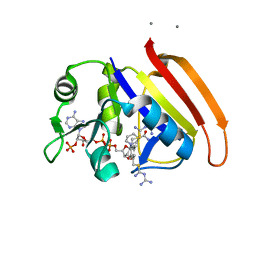 | | Crystal Structure of E.coli DHFR in complex with NADPH and the inhibitor compound 10a. | | Descriptor: | (2,5-dimethylbenzene-1,4-diyl)dimethanediyl bis(N-carbamimidoylcarbamimidothioate), Dihydrofolate reductase, MANGANESE (II) ION, ... | | Authors: | Summerfield, R.L, Daigle, D.M, Mayer, S, Jackson, S.G, Organ, M, Hughes, D.W, Brown, E.D, Junop, M.S. | | Deposit date: | 2005-08-11 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A 2.13 A Structure of E. coli Dihydrofolate Reductase Bound to a Novel Competitive Inhibitor Reveals a New Binding Surface Involving the M20 Loop Region
J.Med.Chem., 49, 2006
|
|
4A45
 
 | | CpGH89CBM32-5, from Clostridium perfringens, in complex with GalNAc- beta-1,3-galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-beta-D-galactopyranose, ALPHA-N-ACETYLGLUCOSAMINIDASE FAMILY PROTEIN, CALCIUM ION, ... | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-10-06 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Carbohydrate Recognition by an Architecturally Complex Alpha-N-Acetylglucosaminidase from Clostridium Perfringens.
Plos One, 7, 2012
|
|
2BQU
 
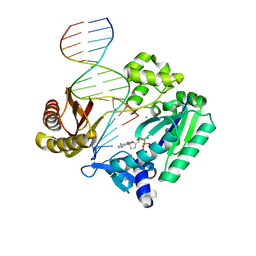 | | DNA Adduct Bypass Polymerization by Sulfolobus solfataricus Dpo4. Analysis and Crystal Structures of Multiple Base-Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*C)-3', 5'-D(*TP*CP*AP*TP*GNEP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2005-04-28 | | Release date: | 2005-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA Adduct Bypass Polymerization by Sulfolobus Solfataricus DNA Polymerase Dpo4: Analysis and Crystal Structures of Multiple Base Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine.
J.Biol.Chem., 280, 2005
|
|
2RAA
 
 | |
3UM6
 
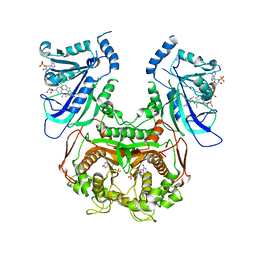 | | Double mutant (A16V+S108T) Plasmodium falciparum DHFR-TS (T9/94) complexed with cycloguanil, NADPH and dUMP | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Chitnumsub, P, Kamchonwongpaisan, S, Yuthavong, Y. | | Deposit date: | 2011-11-12 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Combined Spatial Limitation around Residues 16 and 108 of Plasmodium falciparum Dihydrofolate Reductase Explains Resistance to Cycloguanil.
Antimicrob.Agents Chemother., 56, 2012
|
|
3GYF
 
 | | Human DHFR with Z-isomer in Orthorhombic lattice | | Descriptor: | 5-[(1Z)-2-(2-methoxyphenyl)prop-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V. | | Deposit date: | 2009-04-03 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Z isomer of 2,4-diaminofuro[2,3-d]pyrimidine antifolate promotes unusual crystal packing in a human dihydrofolate reductase ternary complex.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1SXB
 
 | | CRYSTAL STRUCTURE OF REDUCED BOVINE ERYTHROCYTE SUPEROXIDE DISMUTASE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Rypniewski, W.R, Mangani, S, Bruni, B, Orioli, P, Casati, M, Wilson, K.S. | | Deposit date: | 1995-03-17 | | Release date: | 1995-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of reduced bovine erythrocyte superoxide dismutase at 1.9 A resolution.
J.Mol.Biol., 251, 1995
|
|
2GD1
 
 | |
2AZN
 
 | | X-RAY Structure of 2,5-diamino-6-ribosylamino-4(3h)-pyrimidinone 5-phosphate reductase | | Descriptor: | 2-(6-(2-CYCLOHEXYLETHOXY)-TETRAHYDRO-4,5-DIHYDROXY-2(HYDROXYMETHYL)-2H-PYRAN-3-YLOXY)-TETRAHYDRO-6(HYDROXYMETHYL)-2H-PY RAN-3,4,5-TRIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Chatwell, L, Bacher, A, Huber, R, Fischer, M, Krojer, T. | | Deposit date: | 2005-09-12 | | Release date: | 2006-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biosynthesis of riboflavin: structure and properties of 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5'-phosphate reductase of Methanocaldococcus jannaschii
J.Mol.Biol., 359, 2006
|
|
2RBW
 
 | | Cytochrome C Peroxidase W191G in complex with 1,2-dimethyl-1h-pyridin-5-amine | | Descriptor: | 5-amino-1,2-dimethylpyridinium, Cytochrome C Peroxidase, PHOSPHATE ION, ... | | Authors: | Graves, A.P, Boyce, S.E, Shoichet, B.K. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rescoring docking hit lists for model cavity sites: predictions and experimental testing.
J.Mol.Biol., 377, 2008
|
|
4AJN
 
 | | rat LDHA in complex with 2-((4-(2-((3-((2-methyl-1,3-benzothiazol-6- yl)amino)-3-oxo-propyl)carbamoylamino)ethyl)phenyl)methyl) propanedioic acid | | Descriptor: | (4-{2-[({3-[(2-METHYL-1,3-BENZOTHIAZOL-6-YL)AMINO]-3-OXOPROPYL}CARBAMOYL)AMINO]ETHYL}BENZYL)PROPANEDIOIC ACID, GLYCEROL, L-LACTATE DEHYDROGENASE A CHAIN | | Authors: | Tucker, J.A, Brassington, C, Hassall, G, Watson, M, Ward, R, Tart, J, Davies, G, Caputo, A, Pearson, S. | | Deposit date: | 2012-02-16 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Design and Synthesis of Novel Lactate Dehydrogenase a Inhibitors by Fragment-Based Lead Generation
J.Med.Chem., 55, 2012
|
|
3E4V
 
 | |
3NU0
 
 | | Design, Synthesis, Biological Evaluation and X-ray Crystal Structure of Novel Classical 6,5,6-TricyclicBenzo[4,5]thieno[2,3-d]pyrimidines as Dual Thymidylate Synthase and Dihydrofolate Reductase Inhibitors | | Descriptor: | (2S)-2-(5-{[(2-amino-4-oxo-3,4-dihydro[1]benzothieno[2,3-d]pyrimidin-5-yl)methyl]amino}-1-oxo-1,3-dihydro-2H-isoindol-2-yl)pentanedioic acid, Dihydrofolate reducatase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V. | | Deposit date: | 2010-07-06 | | Release date: | 2011-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray crystal structure of novel classical 6,5,6-tricyclic benzo[4,5]thieno[2,3-d]pyrimidines as dual thymidylate synthase and dihydrofolate reductase inhibitors.
Bioorg.Med.Chem., 19, 2011
|
|
2BQR
 
 | | DNA Adduct Bypass Polymerization by Sulfolobus solfataricus Dpo4. Analysis and Crystal Structures of Multiple Base-Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*C)-3', 5'-D(*TP*CP*AP*TP*GNEP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2005-04-27 | | Release date: | 2005-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | DNA Adduct Bypass Polymerization by Sulfolobus Solfataricus DNA Polymerase Dpo4: Analysis and Crystal Structures of Multiple Base Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine.
J.Biol.Chem., 280, 2005
|
|
5T0V
 
 | | Architecture of the Yeast Mitochondrial Iron-Sulfur Cluster Assembly Machinery: the Sub-Complex Formed by the Iron Donor, Yfh1, and the Scaffold, Isu1 | | Descriptor: | Frataxin homolog, mitochondrial, Iron sulfur cluster assembly protein 1 | | Authors: | Ranatunga, W, Gakh, O, Galeano, B.K, Smith IV, D.Y, Soderberg, C.A, Al-Karadaghi, S, Thompson, J.R, Isaya, G. | | Deposit date: | 2016-08-16 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (17.5 Å) | | Cite: | Architecture of the Yeast Mitochondrial Iron-Sulfur Cluster Assembly Machinery: the Sub-Complex Formed by the Iron Donor, Yfh1, and the Scaffold, Isu1
J. Biol. Chem., 291, 2016
|
|
