8RU2
 
 | | Structure of the F-actin barbed end bound by formin mDia1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Molecular mechanism of actin filament elongation by formins.
Science, 384, 2024
|
|
1CAA
 
 | | X-RAY CRYSTAL STRUCTURES OF THE OXIDIZED AND REDUCED FORMS OF THE RUBREDOXIN FROM THE MARINE HYPERTHERMOPHILIC ARCHAEBACTERIUM PYROCOCCUS FURIOSUS | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Day, M.W, Hsu, B.T, Joshua-Tor, L, Park, J.B, Zhou, Z.H, Adams, M.W.W, Rees, D.C. | | Deposit date: | 1992-05-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structures of the oxidized and reduced forms of the rubredoxin from the marine hyperthermophilic archaebacterium Pyrococcus furiosus.
Protein Sci., 1, 1992
|
|
1CAH
 
 | |
8RV2
 
 | | Structure of the formin INF2 bound to the barbed end of F-actin. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | Deposit date: | 2024-01-31 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Molecular mechanism of actin filament elongation by formins.
Science, 384, 2024
|
|
2WC7
 
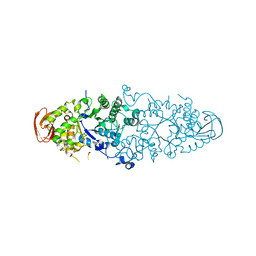 | | Crystal structure of Nostoc Punctiforme Debranching Enzyme(NPDE)(Acarbose soaked) | | Descriptor: | ALPHA AMYLASE, CATALYTIC REGION | | Authors: | Dumbrepatil, A.-B, Song, H.-N, Choi, J.-H, Park, K.-H, Woo, E.-J. | | Deposit date: | 2009-03-10 | | Release date: | 2009-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural Features of the Nostoc Punctiforme Debranching Enzyme Reveal the Basis of its Mechanism and Substrate Specificity.
Proteins, 78, 2010
|
|
1CBG
 
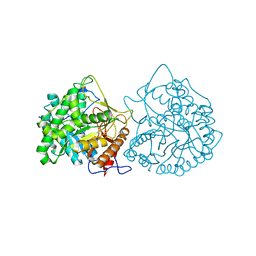 | | THE CRYSTAL STRUCTURE OF A CYANOGENIC BETA-GLUCOSIDASE FROM WHITE CLOVER (TRIFOLIUM REPENS L.), A FAMILY 1 GLYCOSYL-HYDROLASE | | Descriptor: | CYANOGENIC BETA-GLUCOSIDASE | | Authors: | Barrett, T.E, Suresh, C.G, Tolley, S.P, Hughes, M.A. | | Deposit date: | 1995-07-31 | | Release date: | 1995-10-15 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a cyanogenic beta-glucosidase from white clover, a family 1 glycosyl hydrolase.
Structure, 3, 1995
|
|
8Q24
 
 | |
8Q3D
 
 | |
1CCG
 
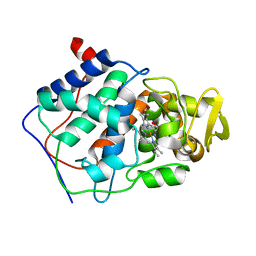 | | CONSTRUCTION OF A BIS-AQUO HEME ENZYME AND REPLACEMENT WITH EXOGENOUS LIGAND | | Descriptor: | CYTOCHROME C PEROXIDASE, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mcree, D.E, Jensen, G.M, Fitzgerald, M.M, Siegel, H.A, Goodin, D.B. | | Deposit date: | 1994-05-04 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Construction of a bisaquo heme enzyme and binding by exogenous ligands.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
8Q26
 
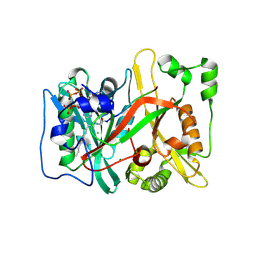 | |
8Q3S
 
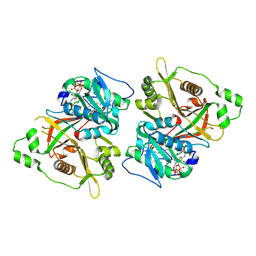 | |
2VF2
 
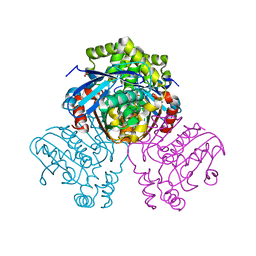 | | X-ray crystal structure of HsaD from Mycobacterium tuberculosis | | Descriptor: | 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, GLYCEROL, SULFATE ION | | Authors: | Lack, N, Lowe, E.D, Liu, J, Eltis, L.D, Noble, M.E.M, Sim, E, Westwood, I.M. | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Hsad, a Steroid-Degrading Hydrolase, from Mycobacterium Tuberculosis.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2XQ8
 
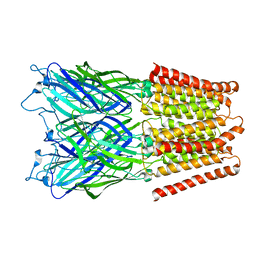 | | Pentameric ligand gated ion channel GLIC in complex with zinc ion (Zn2+) | | Descriptor: | GLR4197 PROTEIN, ZINC ION | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
8Q4F
 
 | | Structure of arbekacin bound Escherichia coli 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 50S ribosomal protein L16, ... | | Authors: | Majumdar, S, Parajuli, N.P, Ge, X, Emmerich, A, Sanyal, S. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of arbekacin bound Escherichia coli 70S ribosome
To be published
|
|
2XQ7
 
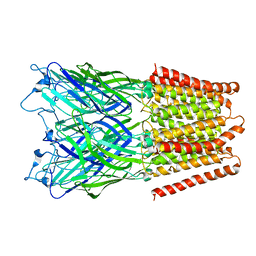 | | Pentameric ligand gated ion channel GLIC in complex with cadmium ion (Cd2+) | | Descriptor: | CADMIUM ION, GLR4197 PROTEIN | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
2V95
 
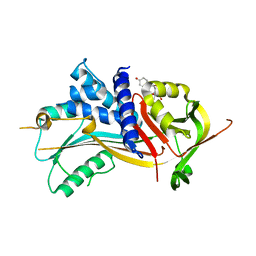 | | Structure of Corticosteroid-Binding Globulin in complex with Cortisol | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, CORTICOSTEROID-BINDING GLOBULIN | | Authors: | Klieber, M.A, Muller, Y.A. | | Deposit date: | 2007-08-21 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Corticosteroid-Binding Globulin: Structural Basis for Steroid Transport and Proteinase-Triggered Release
J.Biol.Chem., 282, 2007
|
|
1CAN
 
 | |
8PZ2
 
 | | Wait Complex: Lateral open BAM bound Extended SurA | | Descriptor: | Chaperone SurA, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Fenn, K.L, Ranson, N.A. | | Deposit date: | 2023-07-26 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Outer membrane protein assembly mediated by BAM-SurA complexes.
Nat Commun, 15, 2024
|
|
8PZU
 
 | |
2XQ5
 
 | | Pentameric ligand gated ion channel GLIC in complex with tetraethylarsonium (TEAs) | | Descriptor: | ARSENIC, GLR4197 PROTEIN | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
1CAO
 
 | |
8PZ1
 
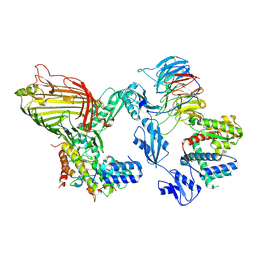 | | Wait Complex: Lateral open BAM bound Compact SurA | | Descriptor: | Chaperone SurA, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Fenn, K.L, Ranson, N.A. | | Deposit date: | 2023-07-26 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Outer membrane protein assembly mediated by BAM-SurA complexes.
Nat Commun, 15, 2024
|
|
2XQ9
 
 | | Pentameric ligand gated ion channel GLIC mutant E221A in complex with tetraethylarsonium (TEAs) | | Descriptor: | ARSENIC, GLR4197 PROTEIN | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
1CCI
 
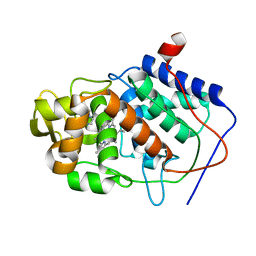 | | HOW FLEXIBLE ARE PROTEINS? TRAPPING OF A FLEXIBLE LOOP | | Descriptor: | 2,3-DIMETHYLIMIDAZOLIUM ION, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cao, Y, Musah, R.A, Wilcox, S.K, Goodin, D.B, Mcree, D.E. | | Deposit date: | 1996-12-18 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A ligand-gated, hinged loop rearrangement opens a channel to a buried artificial protein cavity.
Nat.Struct.Biol., 3, 1996
|
|
8Q0G
 
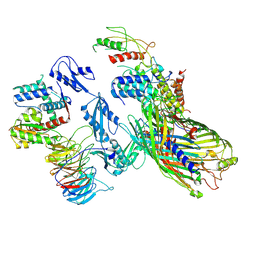 | | Release Complex: BAM bound EspP and Compact SurA | | Descriptor: | Chaperone SurA, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Fenn, K.L, Ranson, N.A. | | Deposit date: | 2023-07-28 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Outer membrane protein assembly mediated by BAM-SurA complexes.
Nat Commun, 15, 2024
|
|
