1UWN
 
 | |
1UZX
 
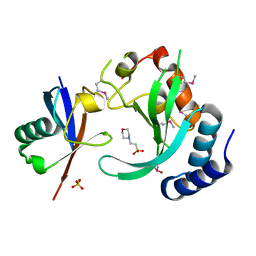 | | A complex of the Vps23 UEV with ubiquitin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, UBIQUITIN, ... | | Authors: | Teo, H, Williams, R.L. | | Deposit date: | 2004-03-18 | | Release date: | 2004-03-30 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights Into Endosomal Sorting Complex Required for Transport (Escrt-I) Recognition of Ubiquitinated Proteins
J.Biol.Chem., 279, 2004
|
|
1UIP
 
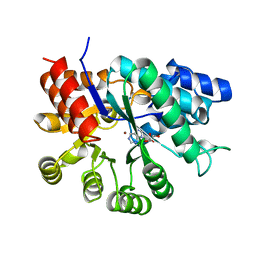 | | ADENOSINE DEAMINASE (HIS 238 GLU MUTANT) | | Descriptor: | ADENOSINE DEAMINASE, PURINE RIBOSIDE, ZINC ION | | Authors: | Wilson, D.K, Quiocho, F.A. | | Deposit date: | 1996-08-30 | | Release date: | 1997-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Site-directed mutagenesis of histidine 238 in mouse adenosine deaminase: substitution of histidine 238 does not impede hydroxylate formation.
Biochemistry, 35, 1996
|
|
1L55
 
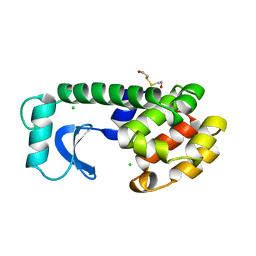 | |
1L0S
 
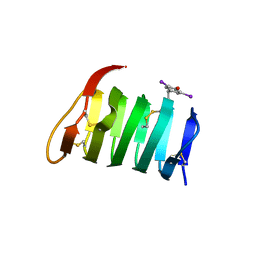 | |
1UMI
 
 | | Structural basis of sugar-recognizing ubiquitin ligase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, F-box only protein 2 | | Authors: | Mizushima, T, Hirao, T, Yoshida, Y, Lee, S.J, Chiba, T, Iwai, K, Yamaguchi, Y, Kato, K, Tsukihara, T, Tanaka, K. | | Deposit date: | 2003-10-01 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of sugar-recognizing ubiquitin ligase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1RNG
 
 | |
1LJL
 
 | | Wild Type pI258 S. aureus arsenate reductase | | Descriptor: | POTASSIUM ION, arsenate reductase | | Authors: | Messens, J, Martins, J.C, Van Belle, K, Brosens, E, Desmyter, A, De Gieter, M, Wieruszeski, J.M, Willem, R, Wyns, L, Zegers, I. | | Deposit date: | 2002-04-21 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | All intermediates of the arsenate reductase mechanism, including an intramolecular dynamic disulfide cascade.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1K9B
 
 | | Crystal structure of the bifunctional soybean Bowman-Birk inhibitor at 0.28 nm resolution. Structural peculiarities in a folded protein conformation | | Descriptor: | BOWMAN-BIRK TYPE PROTEINASE INHIBITOR | | Authors: | Voss, R.H, Ermler, U, Essen, L.O, Wenzl, G, Kim, Y.M, Flecker, P. | | Deposit date: | 2001-10-29 | | Release date: | 2001-11-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the bifunctional soybean Bowman-Birk inhibitor at 0.28-nm resolution. Structural peculiarities in a folded protein conformation.
Eur.J.Biochem., 242, 1996
|
|
1UVC
 
 | | Lipid Binding in Rice Nonspecific Lipid Transfer Protein-1 Complexes from Oryza sativa | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN, STEARIC ACID | | Authors: | Cheng, H.-C, Cheng, P.-T, Peng, P, Lyu, P.-C, Sun, Y.-J. | | Deposit date: | 2004-01-19 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lipid Binding in Rice Nonspecific Lipid Transfer Protein-1 Complexes from Oryza Sativa
Protein Sci., 13, 2004
|
|
1K61
 
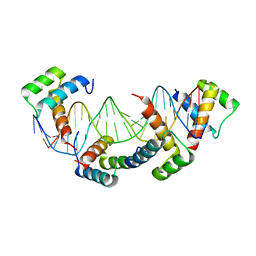 | | MATALPHA2 HOMEODOMAIN BOUND TO DNA | | Descriptor: | 5'-D(*(5IU)P*GP*CP*GP*TP*GP*TP*AP*AP*AP*TP*GP*AP*AP*TP*TP*AP*CP*AP*TP*G)-3', 5'-D(*AP*CP*AP*TP*GP*TP*AP*AP*TP*TP*CP*AP*TP*TP*TP*AP*CP*AP*CP*GP*C)-3', Mating-type protein alpha-2 | | Authors: | Aishima, J, Gitti, R.K, Noah, J.E, Gan, H.H, Schlick, T, Wolberger, C. | | Deposit date: | 2001-10-14 | | Release date: | 2002-12-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Hoogsteen base pair embedded in undistorted B-DNA
NUCLEIC ACIDS RES., 30, 2002
|
|
1UTR
 
 | | UTEROGLOBIN-PCB COMPLEX (REDUCED FORM) | | Descriptor: | 4,4'-BIS([H]METHYLSULFONYL)-2,2',5,5'-TETRACHLOROBIPHENYL, UTEROGLOBIN | | Authors: | Hard, T, Barnes, H.J, Larsson, C, Gustafsson, J.-A, Lund, J. | | Deposit date: | 1995-09-01 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a mammalian PCB-binding protein in complex with a PCB.
Nat.Struct.Biol., 2, 1995
|
|
3HOH
 
 | | RIBONUCLEASE T1 (THR93GLN MUTANT) COMPLEXED WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Langhorst, U, Loris, R, Denisov, V.P, Doumen, J, Roose, P, Maes, D, Halle, B, Steyaert, J. | | Deposit date: | 1998-09-11 | | Release date: | 1998-09-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dissection of the structural and functional role of a conserved hydration site in RNase T1.
Protein Sci., 8, 1999
|
|
1L0Q
 
 | | Tandem YVTN beta-propeller and PKD domains from an archaeal surface layer protein | | Descriptor: | Surface layer protein | | Authors: | Jing, H, Takagi, J, Liu, J.-H, Lindgren, S, Zhang, R.-G, Joachimiak, A, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-02-12 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Archaeal Surface Layer Proteins Contain beta Propeller, PKD, and beta Helix Domains and Are Related to Metazoan Cell Surface Proteins.
Structure, 10, 2002
|
|
1L54
 
 | |
1L61
 
 | |
1V1T
 
 | | Crystal structure of the PDZ tandem of human syntenin in complex with TNEYKV peptide | | Descriptor: | BENZOIC ACID, SYNTENIN 1, TNEYKV PEPTIDE | | Authors: | Grembecka, J, Cierpicki, T, Devedjiev, Y, Cooper, D.R, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2004-04-23 | | Release date: | 2005-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Binding of the Pdz Tandem of Syntenin to Target Proteins.
Biochemistry, 45, 2006
|
|
1KTJ
 
 | | X-ray Structure Of Der P 2, The Major House Dust Mite Allergen | | Descriptor: | ALLERGEN DER P 2 | | Authors: | Derewenda, U, Li, J, Derewenda, Z, Dauter, Z, Mueller, G.A, Rule, G.S, Benjamin, D.C. | | Deposit date: | 2002-01-16 | | Release date: | 2002-05-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a major dust mite allergen Der p 2, and its biological implications.
J.Mol.Biol., 318, 2002
|
|
1V7F
 
 | | Solution structure of phrixotoxin 1 | | Descriptor: | Phrixotoxin 1 | | Authors: | Chagot, B, Escoubas, P, Villegas, E, Bernard, C, Ferrat, G, Corzo, G, Lazdunski, M, Darbon, H. | | Deposit date: | 2003-12-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Phrixotoxin 1, a specific peptide inhibitor of Kv4 potassium channels from the venom of the theraphosid spider Phrixotrichus auratus
Protein Sci., 13, 2004
|
|
1LP1
 
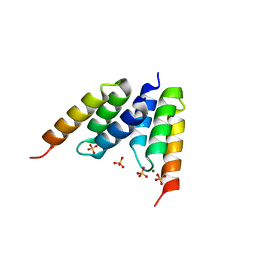 | | Protein Z in complex with an in vitro selected affibody | | Descriptor: | Affibody binding protein Z, Immunoglobulin G binding protein A, MAGNESIUM ION, ... | | Authors: | Hogbom, M, Eklund, M, Nygren, P.A, Nordlund, P. | | Deposit date: | 2002-05-07 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition by an in vitro evolved affibody.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1UUH
 
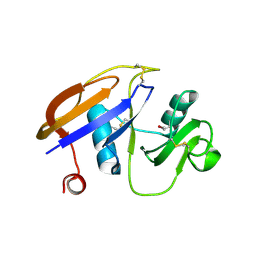 | | Hyaluronan binding domain of human CD44 | | Descriptor: | CD44 ANTIGEN | | Authors: | Teriete, P, Banerji, S, Noble, M, Blundell, C, Wright, A, Pickford, A, Lowe, E, Mahoney, D, Tammi, M, Kahmann, J, Campbell, I, Day, A, Jackson, D. | | Deposit date: | 2003-12-19 | | Release date: | 2004-03-04 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Regulatory Hyaluronan-Binding Domain in the Inflammatory Leukocyte Homing Receptor Cd44
Mol.Cell, 13, 2004
|
|
1UMH
 
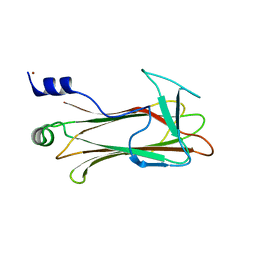 | | Structural basis of sugar-recognizing ubiquitin ligase | | Descriptor: | F-box only protein 2, NICKEL (II) ION | | Authors: | Mizushima, T, Hirao, T, Yoshida, Y, Lee, S.J, Chiba, T, Iwai, K, Yamaguchi, Y, Kato, K, Tsukihara, T, Tanaka, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-01 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of sugar-recognizing ubiquitin ligase
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
1T85
 
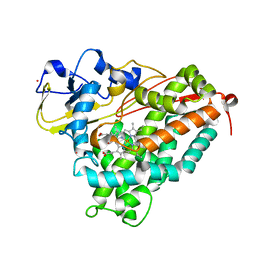 | | Crystal Structure of the Ferrous CO-bound Cytochrome P450cam Mutant (L358P/C334A) | | Descriptor: | CAMPHOR, CARBON MONOXIDE, Cytochrome P450-cam, ... | | Authors: | Nagano, S, Tosha, T, Ishimori, K, Morishima, I, Poulos, T.L. | | Deposit date: | 2004-05-11 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the cytochrome p450cam mutant that exhibits the same spectral perturbations induced by putidaredoxin binding.
J.Biol.Chem., 279, 2004
|
|
1T28
 
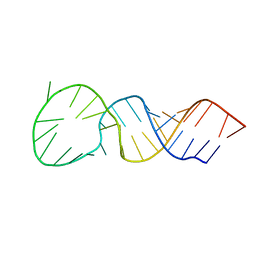 | | High resolution structure of a picornaviral internal cis-acting replication element | | Descriptor: | 34-MER | | Authors: | Thiviyanathan, V, Yang, Y, Kaluarachchi, K, Reynbrand, R, Gorenstein, D.G, Lemon, S.M. | | Deposit date: | 2004-04-20 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution structure of a picornaviral internal cis-acting replication element(cre).
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1UN3
 
 | |
