1XGS
 
 | |
1XGO
 
 | |
1XAC
 
 | |
1XYN
 
 | |
1XAD
 
 | |
1XRC
 
 | | CRYSTAL STRUCTURE OF S-ADENOSYLMETHIONINE SYNTHETASE | | Descriptor: | COBALT (II) ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Takusagawa, F, Kamitori, S, Misaki, S, Markham, G.D. | | Deposit date: | 1995-10-26 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of S-adenosylmethionine synthetase.
J.Biol.Chem., 271, 1996
|
|
2FBX
 
 | | WRN exonuclease, Mg complex | | Descriptor: | MAGNESIUM ION, Werner syndrome helicase | | Authors: | Perry, J.J. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
1XVA
 
 | | METHYLTRANSFERASE | | Descriptor: | ACETATE ION, GLYCINE N-METHYLTRANSFERASE, S-ADENOSYLMETHIONINE | | Authors: | Fu, Z, Hu, Y, Konishi, K, Takata, Y, Ogawa, H, Gomi, T, Fujioka, M, Takusagawa, F. | | Deposit date: | 1996-07-20 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glycine N-methyltransferase from rat liver.
Biochemistry, 35, 1996
|
|
1XAB
 
 | |
1XAA
 
 | |
1XGM
 
 | |
1XRA
 
 | | CRYSTAL STRUCTURE OF S-ADENOSYLMETHIONINE SYNTHETASE | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Takusagawa, F, Kamitori, S, Misaki, S, Markham, G.D. | | Deposit date: | 1995-10-26 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of S-adenosylmethionine synthetase.
J.Biol.Chem., 271, 1996
|
|
1XXC
 
 | |
1XPT
 
 | | BOVINE RIBONUCLEASE A (PHOSPHATE-FREE) | | Descriptor: | RIBONUCLEASE A | | Authors: | Sadasivan, C, Nagendra, H.G, Vijayan, M. | | Deposit date: | 1998-02-23 | | Release date: | 1998-05-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Plasticity, hydration and accessibility in ribonuclease A. The structure of a new crystal form and its low-humidity variant.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1XPS
 
 | |
1XPB
 
 | | STRUCTURE OF BETA-LACTAMASE TEM1 | | Descriptor: | BETA-LACTAMASE, SULFATE ION | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 1997-01-10 | | Release date: | 1997-04-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | TEM1 beta-lactamase structure solved by molecular replacement and refined structure of the S235A mutant.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1XRB
 
 | | S-adenosylmethionine synthetase (MAT, ATP: L-methionine S-adenosyltransferase, E.C.2.5.1.6) in which MET residues are replaced with selenomethionine residues (MSE) | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Takusagawa, F, Kamitori, S, Misaki, S, Markham, G.D. | | Deposit date: | 1995-10-26 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of S-adenosylmethionine synthetase.
J.Biol.Chem., 271, 1996
|
|
1XUG
 
 | | TRYPSIN-BABIM-ZN+2, PH 8.2 | | Descriptor: | BIS(5-AMIDINO-BENZIMIDAZOLYL)METHANE, CALCIUM ION, TRYPSIN, ... | | Authors: | Katz, B.A, Clark, J.M, Finer-Moore, J.S, Jenkins, T.E, Johnson, C.R, Rose, M.J, Luong, C, Moore, W.R, Stroud, R.M. | | Deposit date: | 1997-10-10 | | Release date: | 1998-12-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of potent selective zinc-mediated serine protease inhibitors.
Nature, 391, 1998
|
|
1YEF
 
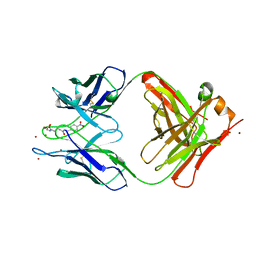 | | STRUCTURE OF IGG2A FAB FRAGMENT (D2.3) COMPLEXED WITH SUBSTRATE ANALOGUE | | Descriptor: | IGG2A FAB FRAGMENT, PARA-NITROBENZYL GLUTARYL GLYCINIC ACID, ZINC ION | | Authors: | Gigant, B, Knossow, M. | | Deposit date: | 1997-05-29 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of a hydrolytic antibody and of complexes elucidate catalytic pathway from substrate binding and transition state stabilization through water attack and product release.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1YDC
 
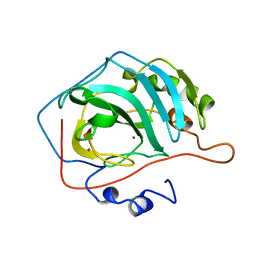 | |
1YE1
 
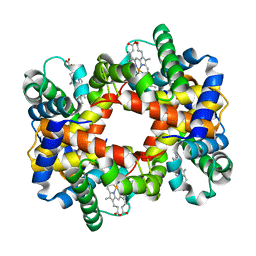 | | T-To-T(High) quaternary transitions in human hemoglobin: betaY35A oxy (2MM IHP, 20% PEG) (1 test set) | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kavanaugh, J.S, Rogers, P.H, Arnone, A. | | Deposit date: | 2004-12-27 | | Release date: | 2005-01-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Crystallographic evidence for a new ensemble of ligand-induced allosteric transitions in hemoglobin: the T-to-T(high) quaternary transitions.
Biochemistry, 44, 2005
|
|
1YEJ
 
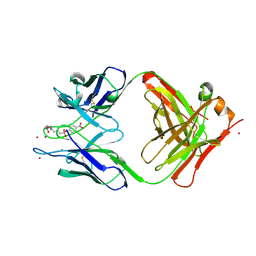 | | CATALYTIC ANTIBODY COMPLEX | | Descriptor: | 6-{4-[HYDROXY-(4-NITRO-PHENOXY)-PHOSPHORYL]-BUTYRYLAMINO}-HEXANOIC ACID, PROTEIN (IG ANTIBODY D2.3 (HEAVY CHAIN)), PROTEIN (IG ANTIBODY D2.3 (LIGHT CHAIN)), ... | | Authors: | Gigant, B, Knossow, M. | | Deposit date: | 1998-08-13 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crossreactivity, efficiency and catalytic specificity of an esterase-like antibody.
J.Mol.Biol., 284, 1998
|
|
1Y2G
 
 | | Crystal STructure of ZipA in complex with an inhibitor | | Descriptor: | Cell division protein zipA, N-METHYL-N-[3-(6-PHENYL[1,2,4]TRIAZOLO[4,3-B]PYRIDAZIN-3-YL)PHENYL]ACETAMIDE | | Authors: | Mosyak, L, Rush, T.S. | | Deposit date: | 2004-11-22 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A shape-based 3-D scaffold hopping method and its application to a bacterial protein-protein interaction
J.Med.Chem., 48, 2005
|
|
1YGS
 
 | | CRYSTAL STRUCTURE OF THE SMAD4 TUMOR SUPPRESSOR C-TERMINAL DOMAIN | | Descriptor: | SMAD4 | | Authors: | Shi, Y, Hata, A, Lo, R.S, Massague, J, Pavletich, N.P. | | Deposit date: | 1997-10-03 | | Release date: | 1998-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural basis for mutational inactivation of the tumour suppressor Smad4.
Nature, 388, 1997
|
|
1ZIN
 
 | | ADENYLATE KINASE WITH BOUND AP5A | | Descriptor: | ADENYLATE KINASE, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ZINC ION | | Authors: | Berry, M.B, Phillips Jr, G.N. | | Deposit date: | 1996-06-07 | | Release date: | 1997-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of Bacillus stearothermophilus adenylate kinase with bound Ap5A, Mg2+ Ap5A, and Mn2+ Ap5A reveal an intermediate lid position and six coordinate octahedral geometry for bound Mg2+ and Mn2+.
Proteins, 32, 1998
|
|
