5B47
 
 | | 2-Oxoacid:Ferredoxin Oxidoreductase 2 from Sulfolobus tokodai - pyruvate complex | | Descriptor: | 2-oxoacid--ferredoxin oxidoreductase alpha subunit, 2-oxoacid--ferredoxin oxidoreductase beta subunit, IRON/SULFUR CLUSTER, ... | | Authors: | Yan, Z, Maruyama, A, Arakawa, T, Fushinobu, S, Wakagi, T. | | Deposit date: | 2016-04-01 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of archaeal 2-oxoacid:ferredoxin oxidoreductases from Sulfolobus tokodaii
Sci Rep, 6, 2016
|
|
5XB8
 
 | | Crystal structure of dibenzothiophene monooxygenase (TdsC) from Paenibacillus sp. A11-2 | | Descriptor: | SULFATE ION, Thermophilic dibenzothiophene desulfurization enzyme C | | Authors: | Hino, T, Hamamoto, H, Ohshiro, T, Nagano, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity.
J. Biol. Chem., 292, 2017
|
|
8C3N
 
 | |
8HOU
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A-T268V in complex with Im-N-C4-Phe-Phe | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-N-C4-Phe-Phe, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8OXT
 
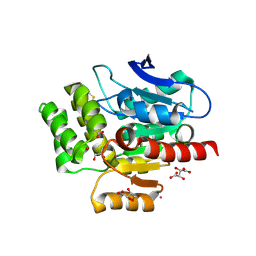 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) H251A VARIANT COMPLEXED WITH N-ACETYLANTHRANILATE AS RESULT OF IN CRYSTALLO TURNOVER OF ITS NATURAL SUBSTRATE 1-H-3-HYDROXY-4- OXOQUINALDINE UNDER HYPEROXIC CONDITIONS | | Descriptor: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, 2-(ACETYLAMINO)BENZOIC ACID, GLYCEROL, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2023-05-02 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
8HOT
 
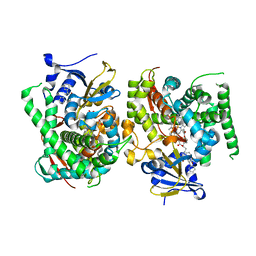 | |
8HOP
 
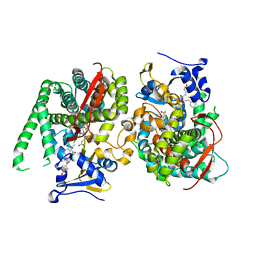 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Nap-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Nap-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5XDD
 
 | | Crystal structure of tertiary complex of TdsC from Paenibacillus sp. A11-2 with FMN and Indole | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, INDOLE, ... | | Authors: | Hino, T, Hamamoto, H, Ohshiro, T, Nagano, S. | | Deposit date: | 2017-03-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity.
J. Biol. Chem., 292, 2017
|
|
8HON
 
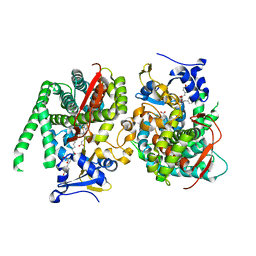 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Tyr-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Tyr-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6UNN
 
 | |
6TUS
 
 | | human XPG, Apo2 form | | Descriptor: | DNA repair protein complementing XP-G cells,DNA repair protein complementing XP-G cells, GLYCEROL, SULFATE ION | | Authors: | Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2020-01-08 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of human XPG, the xeroderma pigmentosum group G endonuclease, provides insight into nucleotide excision DNA repair.
Nucleic Acids Res., 48, 2020
|
|
5B7P
 
 | |
8OPV
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Resveratrol (Fragment-B-H11) | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OQU
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-92 | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, 4-chloranylbenzenesulfonic acid, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OVB
 
 | |
8PDD
 
 | | Thioredoxin glutathione reductase of Schistosoma mansoni at 1.25A resolution. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ribeiro, L, Montoya, B.O, Moreira-Filho, J.T, Bowyer, S, Verma, A, Neves, B.J, Owens, R.J, Andrade, C.H, Silva-Jr, F.P, Furnham, N. | | Deposit date: | 2023-06-12 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Fragment library screening by X-ray crystallography and binding site analysis on thioredoxin glutathione reductase of Schistosoma mansoni.
Sci Rep, 14, 2024
|
|
6TVI
 
 | | Salmonella typhimurium mutant neuraminidase (D100S)+ DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, Sialidase | | Authors: | Garman, E.F, Salinger, M.T, Murray, J.W, Laver, W.G, Kuhn, P, Vimr, E.R. | | Deposit date: | 2020-01-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Salmonella typhimurium mutant neuraminidase (D100S)+ DANA
To Be Published
|
|
8HPG
 
 | |
6TW4
 
 | | HumRadA22F in complex with compound 6 | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA, ~{N}-[2-[(2~{S})-2-[[(1~{S})-1-(4-methoxyphenyl)ethyl]carbamoyl]pyrrolidin-1-yl]-2-oxidanylidene-ethyl]quinoline-2-carboxamide | | Authors: | Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
1IFS
 
 | | RICIN A-CHAIN (RECOMBINANT) COMPLEX WITH ADENOSINE (ADENOSINE BECOMES ADENINE IN THE COMPLEX) | | Descriptor: | ADENINE, RICIN | | Authors: | Weston, S.A, Tucker, A.D, Thatcher, D.R, Derbyshire, D.J, Pauptit, R.A. | | Deposit date: | 1996-07-05 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of recombinant ricin A-chain at 1.8 A resolution.
J.Mol.Biol., 244, 1994
|
|
6TW9
 
 | | HumRadA22F in complex with CAM833 | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA, GLYCEROL, ... | | Authors: | Fischer, G, Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
6TWK
 
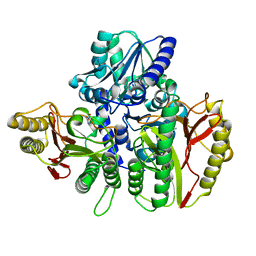 | | Substrate bound structure of the Ectoine utilization protein EutD (DoeA) from Halomonas elongata | | Descriptor: | (2~{R})-4-azanyl-2-[[(1~{S})-1-oxidanylethyl]amino]butanoic acid, (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Ectoine hydrolase DoeA | | Authors: | Mais, C.-N, Altegoer, F, Bange, G. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Degradation of the microbial stress protectants and chemical chaperones ectoine and hydroxyectoine by a bacterial hydrolase-deacetylase complex.
J.Biol.Chem., 295, 2020
|
|
8HT0
 
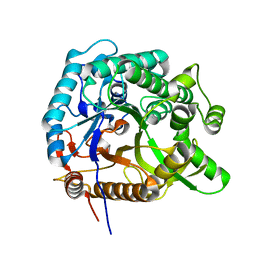 | |
5IIU
 
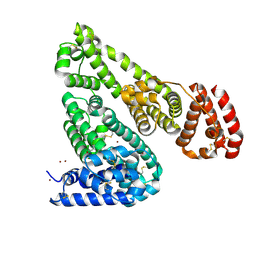 | | Crystal structure of Equine Serum Albumin in the presence of 10 mM zinc at pH 6.9 | | Descriptor: | SULFATE ION, Serum albumin, ZINC ION | | Authors: | Handing, K.B, Shabalin, I.G, Cooper, D.R, Cymborowski, M.T, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-03-01 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Circulatory zinc transport is controlled by distinct interdomain sites on mammalian albumins.
Chem Sci, 7, 2016
|
|
8P00
 
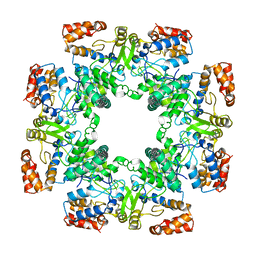 | | Cryo-EM structure of Rotavirus B NSP2 | | Descriptor: | Non-structural protein 2 | | Authors: | Chamera, S, Nowotny, M. | | Deposit date: | 2023-05-09 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of rotavirus B NSP2 reveals its unique tertiary architecture.
J.Virol., 98, 2024
|
|
