3HPG
 
 | |
3ROX
 
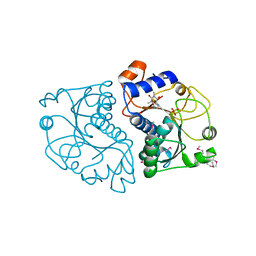 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with Theophylline | | Descriptor: | Apolipoprotein A-I-binding protein, SULFATE ION, THEOPHYLLINE | | Authors: | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2011-04-26 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3R10
 
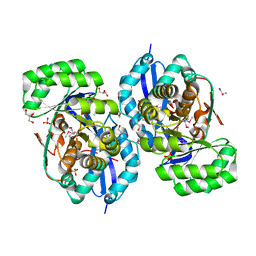 | | Crystal structure of NYSGRC enolase target 200555, a putative dipeptide epimerase from Francisella philomiragia : Mg complex | | Descriptor: | Enzyme of enolase superfamily, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-03-09 | | Release date: | 2011-04-20 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RQ5
 
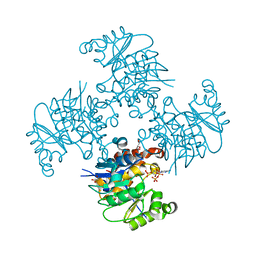 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis co-crystallized with ATP/Mg2+ and soaked with CoA | | Descriptor: | ADP/ATP-dependent NAD(P)H-hydrate dehydratase, COENZYME A, GLYCEROL | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3IUU
 
 | |
3RQH
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis in complex with P1,P6-Di(adenosine-5') hexaphosphate | | Descriptor: | ADP/ATP-DEPENDENT NAD(P)H-HYDRATE DEHYDRATASE, MAGNESIUM ION, P1,P6-Di(adenosine-5') hexaphosphate | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-28 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3IV3
 
 | | The Structure of a putative tagatose 1,6-aldolase from Streptococcus mutans | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Cuff, M.E, Hatzos, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of a putative tagatose 1,6-aldolase from Streptococcus mutans
TO BE PUBLISHED
|
|
3IPT
 
 | |
3IVI
 
 | | Design and Synthesis of Potent BACE-1 Inhibitors with Cellular Activity: Structure-Activity Relationship of P1 Substituents | | Descriptor: | Beta-secretase 1, GLYCEROL, N-[(1S,2R)-3-{[(5S)-5-(3-tert-butylphenyl)-4,5,6,7-tetrahydro-1H-indazol-5-yl]amino}-1-(3,5-difluorobenzyl)-2-hydroxypropyl]acetamide, ... | | Authors: | Pan, H. | | Deposit date: | 2009-09-01 | | Release date: | 2010-01-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and synthesis of cell potent BACE-1 inhibitors: structure-activity relationship of P1' substituents.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3IS5
 
 | | Crystal structure of CDPK kinase domain from toxoplasma Gondii, TGME49_018720 | | Descriptor: | CALCIUM ION, Calcium-dependent protein kinase, GLYCEROL, ... | | Authors: | Wernimont, A.K, Artz, J.D, Senisterra, G, MacKenzie, F, Hutchinson, A, Kozieradzki, I, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-25 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of CDPK kinase domain from toxoplasma Gondii, TGME49_018720
To be Published
|
|
3IW7
 
 | | Human p38 MAP Kinase in Complex with an Imidazo-pyridine | | Descriptor: | 2-({4-[(4-benzylpiperidin-1-yl)carbonyl]benzyl}sulfanyl)-3H-imidazo[4,5-c]pyridine, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Rauh, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-Throughput Screening To Identify Inhibitors Which Stabilize Inactive Kinase Conformations in p38alpha
J.Am.Chem.Soc., 131, 2009
|
|
3R30
 
 | | MK2 kinase bound to Compound 2 | | Descriptor: | 1-(2-aminoethyl)-3-[2-(quinolin-3-yl)pyridin-4-yl]-1H-pyrazole-5-carboxylic acid, MAP kinase-activated protein kinase 2 | | Authors: | Oubrie, A, Fisher, M. | | Deposit date: | 2011-03-15 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-based lead identification of ATP-competitive MK2 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3ITB
 
 | | Crystal structure of Penicillin-Binding Protein 6 (PBP6) from E. coli in complex with a substrate fragment | | Descriptor: | D-alanyl-D-alanine carboxypeptidase DacC, Peptidoglycan substrate (AMV)A(FGA)K(DAL)(DAL), SULFATE ION, ... | | Authors: | Chen, Y, Zhang, W, Shi, Q, Hesek, D, Lee, M, Mobashery, S, Shoichet, B.K. | | Deposit date: | 2009-08-27 | | Release date: | 2009-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of penicillin-binding protein 6 from Escherichia coli.
J.Am.Chem.Soc., 131, 2009
|
|
3RSO
 
 | | H-Ras soaked in 20% S,R,S-bisfuranol: 1 of 10 in MSCS set | | Descriptor: | (3S,3aR,6aS)-hexahydrofuro[2,3-b]furan-3-ol, CALCIUM ION, GTPase HRas, ... | | Authors: | Mattos, C, Buhrman, G, Kearney, B. | | Deposit date: | 2011-05-02 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analysis of Binding Site Hot Spots on the Surface of Ras GTPase.
J.Mol.Biol., 413, 2011
|
|
3R3M
 
 | |
3RSY
 
 | | Cellobiose phosphorylase from Cellulomonas uda in complex with sulfate and glycerol | | Descriptor: | Cellobiose phosphorylase, GLYCEROL, SULFATE ION | | Authors: | Van Hoorebeke, A, Stout, J, Soetaert, W, Van Beeumen, J, Desmet, T, Savvides, S. | | Deposit date: | 2011-05-02 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Cellobiose phosphorylase: reconstructing the structural itinerary along the catalytic pathway
To be Published
|
|
3R40
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/apo | | Descriptor: | CALCIUM ION, CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Chan, P.W.Y, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-03-16 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Mapping the reaction coordinates of enzymatic defluorination.
J.Am.Chem.Soc., 133, 2011
|
|
3IV0
 
 | |
3I9W
 
 | |
3R16
 
 | | Human CAII bound to N-(4-sulfamoylphenyl)-2-(thiophen-2-yl) acetamide | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Biswas, S, McKenna, R, Supuran, C.T. | | Deposit date: | 2011-03-09 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational variability of different sulfonamide inhibitors with thienyl-acetamido moieties attributes to differential binding in the active site of cytosolic human carbonic anhydrase isoforms.
Bioorg.Med.Chem., 19, 2011
|
|
3IW4
 
 | | Crystal structure of PKC alpha in complex with NVP-AEB071 | | Descriptor: | 3-(1H-indol-3-yl)-4-[2-(4-methylpiperazin-1-yl)quinazolin-4-yl]-1H-pyrrole-2,5-dione, Protein kinase C alpha type | | Authors: | Stark, W, Rummel, G, Strauss, A, Cowan-Jacob, S.W. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 3-(1H-indol-3-yl)-4-[2-(4-methylpiperazin-1-yl)quinazolin-4-yl]pyrrole-2,5-dione (AEB071), a potent and selective inhibitor of protein kinase C isotypes
J.Med.Chem., 52, 2009
|
|
3R1L
 
 | | Crystal structure of the Class I ligase ribozyme-substrate preligation complex, C47U mutant, Mg2+ bound | | Descriptor: | 5'-R(*UP*CP*CP*AP*GP*UP*A)-3', Class I ligase ribozyme, MAGNESIUM ION, ... | | Authors: | Shechner, D.M, Bartel, D.P. | | Deposit date: | 2011-03-10 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.125 Å) | | Cite: | The structural basis of RNA-catalyzed RNA polymerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3R2Q
 
 | | Crystal Structure Analysis of yibF from E. Coli | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, PHOSPHATE ION, ... | | Authors: | Ladner, J.E, Harp, J, Schaab, M, Stournan, N.V, Armstrong, R.N. | | Deposit date: | 2011-03-14 | | Release date: | 2012-03-14 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural and Functional Genomics of YibF, a Glutathione Transferase Homologue from Escherichia coli
to be published
|
|
3IXP
 
 | |
3R6Y
 
 | | Crystal structure of chymotrypsin-treated aspartase from Bacillus sp. YM55-1 | | Descriptor: | Aspartase, CALCIUM ION | | Authors: | Fibriansah, G, Puthan Veetil, V, Poelarends, G.J, Thunnissen, A.-M.W.H. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the catalytic mechanism of aspartate ammonia lyase.
Biochemistry, 50, 2011
|
|
