5LP3
 
 | |
7LX8
 
 | | T4 lysozyme mutant L99A | | Descriptor: | 1-chloro-2-(methylsulfanyl)benzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Lysozyme | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-03-03 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5LGE
 
 | | Crystal Structure of human IDH1 mutant (R132H) in complex with NADP+ and an Inhibitor related to BAY 1436032 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(4-propan-2-ylphenyl)amino]-1-[(1~{S},5~{S})-3,3,5-trimethylcyclohexyl]benzimidazole-5-carboxylic acid, ACETATE ION, ... | | Authors: | Hillig, R.C, Hars, U, Korndoerfer, I.P. | | Deposit date: | 2016-07-07 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pan-mutant IDH1 inhibitor BAY 1436032 for effective treatment of IDH1 mutant astrocytoma in vivo.
Acta Neuropathol., 133, 2017
|
|
7LX9
 
 | | T4 lysozyme mutant L99A | | Descriptor: | (but-3-en-1-yl)benzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Lysozyme | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-03-03 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8U3N
 
 | |
7MBX
 
 | | Human Cholecystokinin 1 receptor (CCK1R) Gs complex | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Cholecystokinin receptor type A, Cholecystokinin-8, ... | | Authors: | Mobbs, J.I, Belousoff, M.J, Danev, R, Thal, D.M, Sexton, P.M. | | Deposit date: | 2021-04-01 | | Release date: | 2021-05-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (1.95 Å) | | Cite: | Structures of the human cholecystokinin 1 (CCK1) receptor bound to Gs and Gq mimetic proteins provide insight into mechanisms of G protein selectivity.
Plos Biol., 19, 2021
|
|
9CB5
 
 | | Crystal structure of nucleolin in complex with MYC promoter G-quadruplex | | Descriptor: | Fab heavy chain, Fab light chain, MYC promoter G-quadruplex, ... | | Authors: | Chen, L, Dickerhoff, J, Noinaj, N, Yang, D. | | Deposit date: | 2024-06-18 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for nucleolin recognition of MYC promoter G-quadruplex.
Science, 388, 2025
|
|
8UNT
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (Class I) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8UNY
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (Class N) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
7MBY
 
 | | Human Cholecystokinin 1 receptor (CCK1R) Gq chimera (mGsqi) complex | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Cholecystokinin receptor type A, Cholecystokinin-8, ... | | Authors: | Mobbs, J.I, Belousoff, M.J, Danev, R, Thal, D.M, Sexton, P.M. | | Deposit date: | 2021-04-01 | | Release date: | 2021-05-26 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Structures of the human cholecystokinin 1 (CCK1) receptor bound to Gs and Gq mimetic proteins provide insight into mechanisms of G protein selectivity.
Plos Biol., 19, 2021
|
|
8U7Y
 
 | | Structural Basis of Human NOX5 Activation | | Descriptor: | DECANE, DODECANE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cui, C, Jiang, M, Sun, J. | | Deposit date: | 2023-09-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Structural basis of human NOX5 activation.
Nat Commun, 15, 2024
|
|
1NCC
 
 | | CRYSTAL STRUCTURES OF TWO MUTANT NEURAMINIDASE-ANTIBODY COMPLEXES WITH AMINO ACID SUBSTITUTIONS IN THE INTERFACE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IGG2A-KAPPA NC41 FAB (HEAVY CHAIN), ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of two mutant neuraminidase-antibody complexes with amino acid substitutions in the interface.
J.Mol.Biol., 227, 1992
|
|
8UQV
 
 | |
6NUN
 
 | |
8UQW
 
 | | Round 18 Arylesterase Variant of Apo-Phosphotriesterase Measured at 13 keV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphotriesterase variant PTE-R18 | | Authors: | Breeze, C.W, Frkic, R.L, Campbell, E.C, Jackson, C.J. | | Deposit date: | 2023-10-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mononuclear binding and catalytic activity of europium(III) and gadolinium(III) at the active site of the model metalloenzyme phosphotriesterase.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6NVW
 
 | | Crystal structure of penicillin G acylase from Bacillus megaterium | | Descriptor: | CALCIUM ION, Penicillin G acylase | | Authors: | Blankenfeldt, W. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal structures and protein engineering of three different penicillin G acylases from Gram-positive bacteria with different thermostability.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
7NL2
 
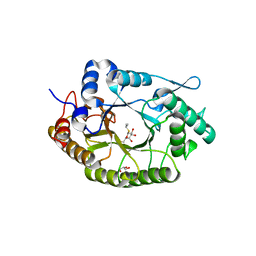 | | Structure of Xyn11 from Pseudothermotoga thermarum | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-xylanase, GLYCEROL | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phylogenetic, functional and structural characterization of a GH10 xylanase active at extreme conditions of temperature and alkalinity
Comput Struct Biotechnol J, 19, 2021
|
|
5LTE
 
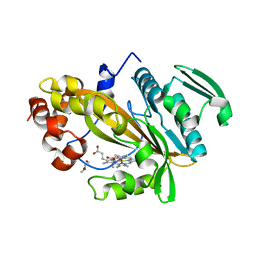 | |
6NZV
 
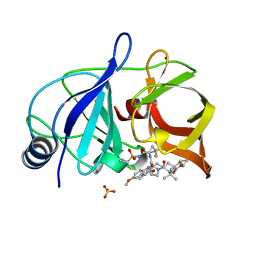 | | Crystal structure of HCV NS3/4A protease in complex with compound 12 | | Descriptor: | (1aR,5S,8S,9S,10R,22aR)-5-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-9-ethyl-14-methoxy-3,6-dioxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadecino[11,12-b]quinoxaline-8-carboxamide, HCV NS3/4A protease, SULFATE ION, ... | | Authors: | Appleby, T.C, Taylor, J.G. | | Deposit date: | 2019-02-14 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of the pan-genotypic hepatitis C virus NS3/4A protease inhibitor voxilaprevir (GS-9857): A component of Vosevi®.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
5LDR
 
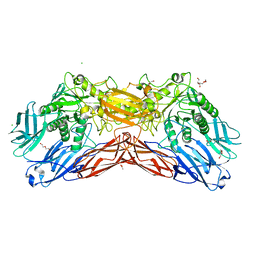 | | Crystal structure of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d strain in complex with galactose | | Descriptor: | ACETATE ION, Beta-D-galactosidase, CHLORIDE ION, ... | | Authors: | Rutkiewicz-Krotewicz, M, Bujacz, A, Pietrzyk, A.J, Sekula, B, Bujacz, G. | | Deposit date: | 2016-06-27 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural studies of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
9CSG
 
 | | Human Serum Albumin Bound to Cerastecin Compound 5e | | Descriptor: | 2-(4-butylbenzene-1-sulfonamido)-5-(4-{3-carboxy-4-[4-(2-methoxyethyl)benzene-1-sulfonamido]anilino}-4-oxobutanamido)benzoic acid, Albumin, MYRISTIC ACID | | Authors: | Hruza, A, Klein, D.J, Ishchenko, A. | | Deposit date: | 2024-07-23 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Cerastecin Inhibition of the Lipooligosaccharide Transporter MsbA to Combat Acinetobacter baumannii : From Screening Impurity to In Vivo Efficacy.
J.Med.Chem., 67, 2024
|
|
5LUS
 
 | | Structures of DHBN domain of Pelecanus crispus BLM helicase | | Descriptor: | BLM helicase | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.433 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
6O18
 
 | |
8UQZ
 
 | | Round 18 Arylesterase Variant of Phosphotriesterase Bound to Gadolinium(III) Measured at 9.5 keV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, GADOLINIUM ION, ... | | Authors: | Breeze, C.W, Frkic, R.L, Campbell, E.C, Jackson, C.J. | | Deposit date: | 2023-10-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Mononuclear binding and catalytic activity of europium(III) and gadolinium(III) at the active site of the model metalloenzyme phosphotriesterase.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6O3I
 
 | | Crystal Structure of Human IDO1 bound to navoximod (NLG-919) | | Descriptor: | Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE, trans-4-{(1R)-2-[(5S)-6-fluoro-5H-imidazo[5,1-a]isoindol-5-yl]-1-hydroxyethyl}cyclohexan-1-ol | | Authors: | Harris, S.F, Oh, A. | | Deposit date: | 2019-02-26 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of Clinical Candidate (1R,4r)-4-((R)-2-((S)-6-Fluoro-5H-imidazo[5,1-a]isoindol-5-yl)-1-hydroxyethyl)cyclohexan-1-ol (Navoximod), a Potent and Selective Inhibitor of Indoleamine 2,3-Dioxygenase 1.
J.Med.Chem., 62, 2019
|
|
